Figure EV1. Differences in curation practices across databases integrated by Pathway Commons.
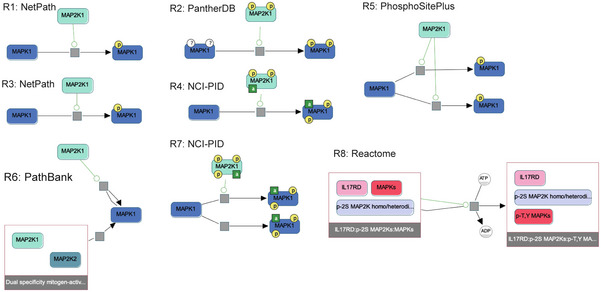
A subgraph of the “paths‐from‐to” query between MAP2K1 and MAPK1 obtained from Pathway Commons and visualized using the ChiBE software (Babur et al, 2009). Each biochemical reaction (R1–R8) depicts a different curation of the same reaction in which MAP2K1 phosphorylates MAPK1. The original source database (e.g., NetPath) is shown next to each reaction. Inconsistencies include (i) the reaction structure itself, with some specifying a single step phosphorylation of two sites (e.g., R4) while others specify single‐site phosphorylation (e.g., R1), or the explicit representation of ADP and ATP as part of the reaction (R8); (ii) the phosphorylation status of MAP2K1, with no phosphorylation status given in R1, R3, R5, R6, and R8, two phosphorylation sites indicated in R2, R4, and three phosphorylation sites in R7; (iii) the initial state of MAPK1, with R2 explicitly indicating unphosphorylated status, while other reactions do not make this explicit; (iv) the final state of MAPK1, with some reactions representing MAPK1 phosphorylation on an unspecified site (R1 and R3), and others providing specific phosphorylation sites (e.g., R2); (v) the specification of active states, with R4 being the only reaction representing MAP2K1 explicitly as active, while R4 and R7 are the only reactions specifying that MAPK1 is active after phosphorylation; and (vi) the presence of other co‐factors such as IL17RD (R8) as part of the reaction.
