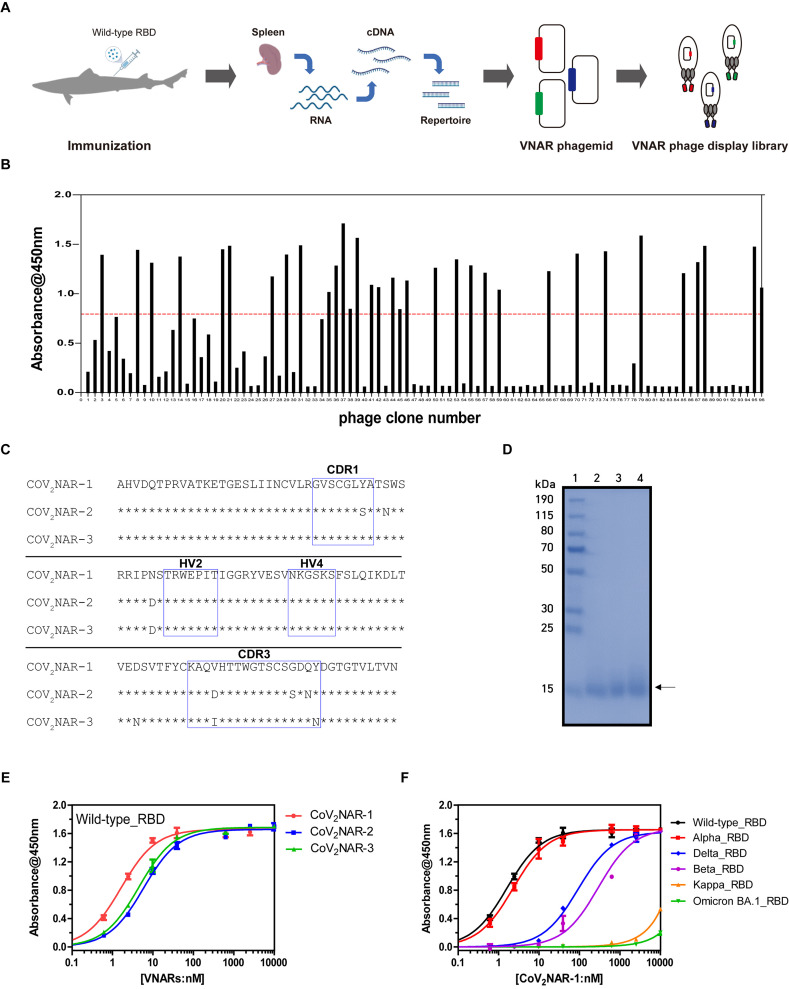
Fig. 1.
Identification of banded houndshark derived VNAR against SARS-CoV-2 RBD. (A) Schematic figure of phage library construction from immunized sharks. (B) Identification of wild-type RBD specific VNAR by the periplasmic extract ELISA. The red dashed line indicates a threshold for the selection of RBD-specific VNARs. (C) Amino acid sequence alignment of CoV2NARs. Asterisks indicate amino acid sequences consistent with CoV2NAR-1. Blue boxes represent CDRs and HVs determined according to the previous reference [24]. (D) SDS-PAGE analysis of purified CoV2NAR-1,2,3. The lanes of SDS-PAGE are as follows: lane 1, protein size marker (Thermo Fisher); lane 2, CoV2NAR-1; lane 3, CoV2NAR-2; and lane 4, CoV2NAR-3. Binding affinity analysis of (E) purified CoV2NARs against the wild-type RBD and (F) CoV2NAR-1 against the RBDs from wild-type and its variants (Alpha, Beta, Delta, Kappa, and Omicron BA.1) by ELISA. Error bars are ±standard deviation for triplicate experiments.