Figure 3(key figure): A model for catalyzed nucleosome-unwrapping facilitating TF-binding.
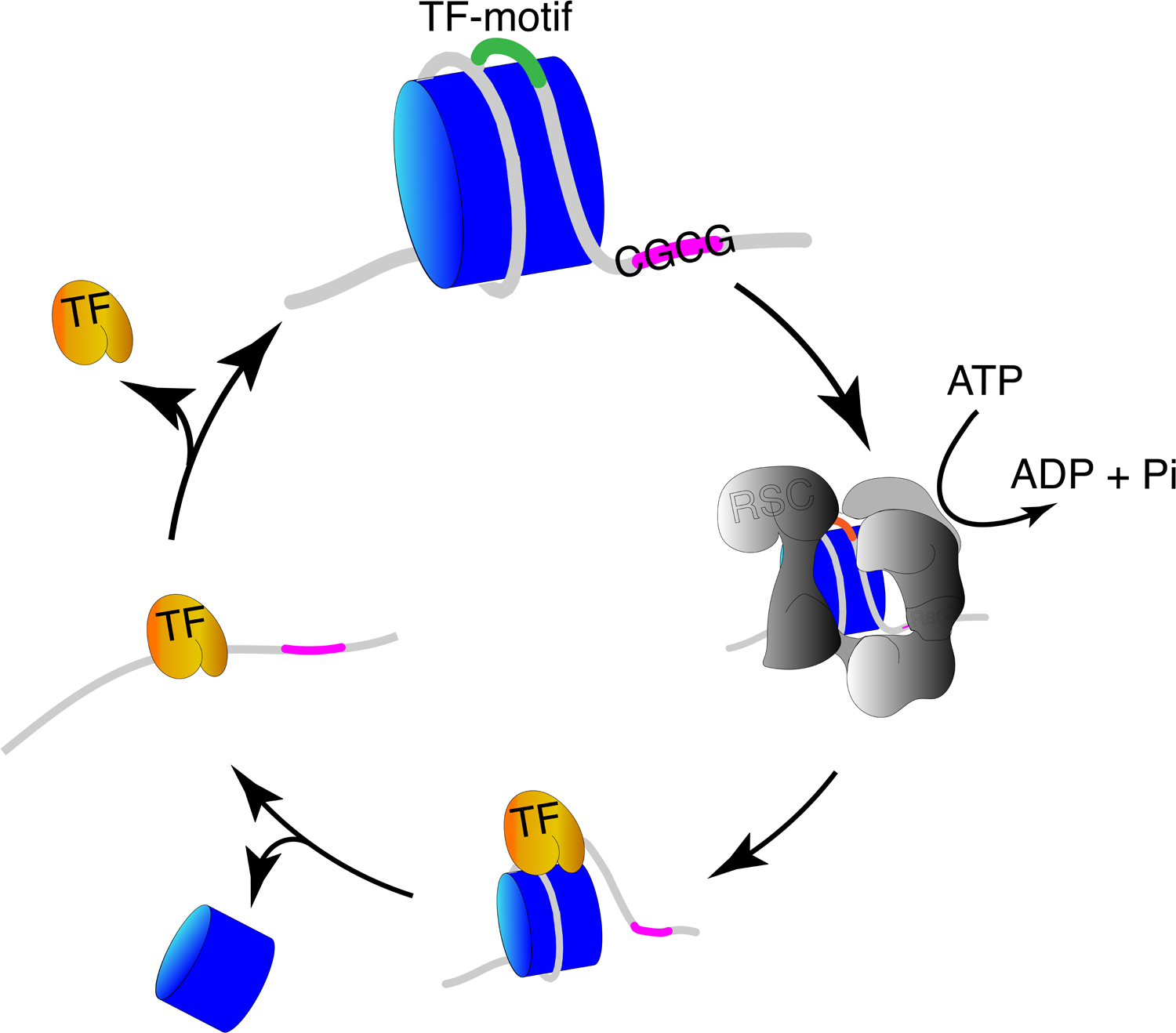
A SWI/SNF remodeler first engulfs a promoter nucleosome behind the replication fork. The remodeler uses energy from ATP hydrolysis to partially unwrap the nucleosome so as to expose the TF-binding motif. TFs (such as the GRFs in yeast) binding to exposed sequence motifs within unwrapped nucleosomes trap the nucleosome in a partially unwrapped state, but the unstable nucleosome is eventually displaced. TFs binding to non-nucleosomal DNA have short dwell-time, and a new nucleosome is deposited, occluding the site for a displaced TF, thus constituting a dynamic cycle.
