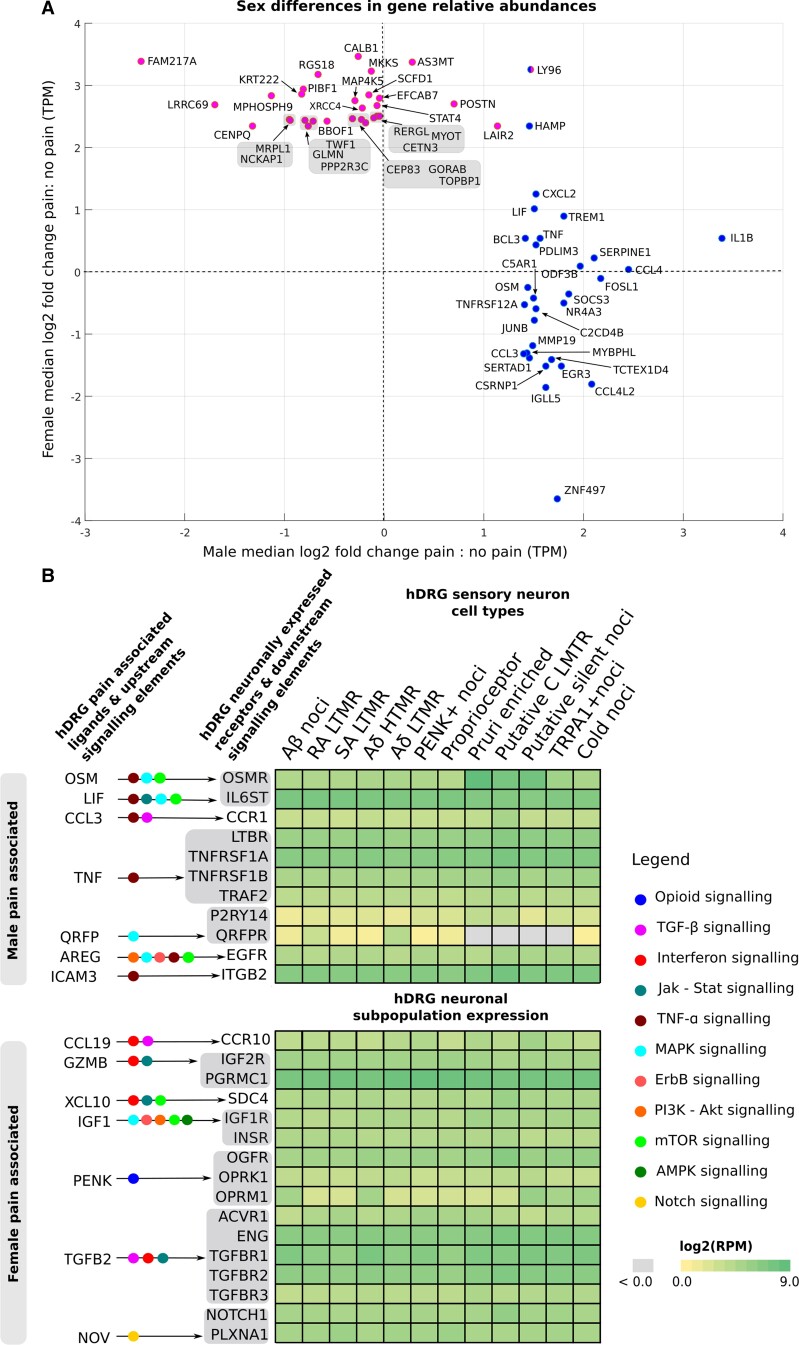
Figure 3.
Sex differential aspects of the pain-associated transcriptome. (A) The top 25 pain-associated genes (Supplementary Table 3A and C) increased in pain (with a median fold change of 2-fold or higher) for each sex show a remarkably sex-differential signal, with only HAMP showing 2-fold or greater change in the median in both sexes among the top 25. Log2 fold changes in the median between pain and no-pain subcohorts are shown for males and females as a scatter plot, with male pain-associated genes shown in blue (right cluster) and female pain-associated genes shown in pink (left cluster). LY96 is present in both male and female lists. (B) Pain-associated ligands in each sex signal to hDRG-expressed receptors that are enriched in sensory neuronal subpopulations,11 but have little overlap across sexes. Based on our interactome analysis, we show such ligand–receptor pairs, alongside receptor expression in human DRG neuronal subtypes11 as a heatmap. Overlap with relevant signalling pathways are also shown. Male signalling is enriched in TNF-alpha pathway, while female signalling is enriched in interferon signalling. Although ICAM3 (in males) and TGFB2 or NOV (in females) are not in the pain-associated gene lists, they are increased in pain for the corresponding sex at the median or upper quartile levels and are thus shown here.