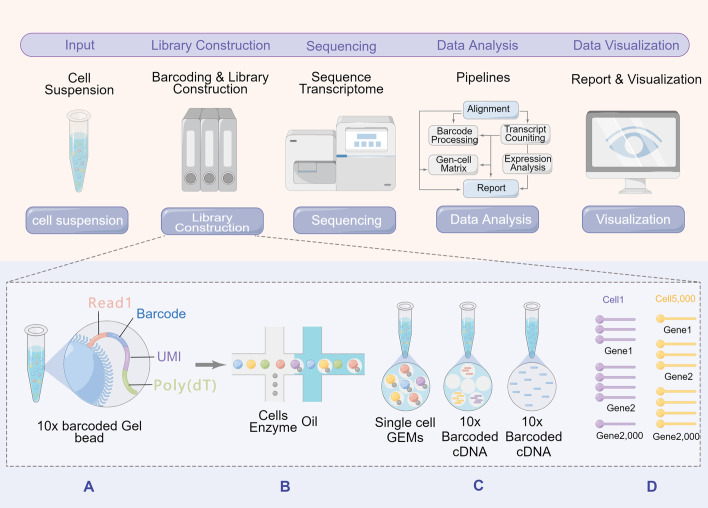
Fig. 1.
Single-cell sequencing flow chart. A Read 1 is derived from beads and carries the sequence of the RNA molecule; Barcode is used to distinguish between cell types; UMI is used to distinguish between RNAs; a UMI is added to each RNA, thus ensuring that the same RNA molecule carries the same UMI after amplification; Poly(dT) on the beads binds to the poly A tail at the end of the RNA. B The cell suspension, beads and oil droplets are added at three locations; when the cell suspension enters, it combines with the beads; after the combination of the two, they are wrapped by the oil droplets, forming an oil-in-water structure. C 10×Barcoded Gel Beads are mixed with cells, enzyme, and partitioning oil. Single cell GEMS undergo reverse transcriptase to generate 10× Barcoded cDNA. All generated cDNA from individual cells shares a common 10× Barcode. D The resulting 10x Barcoded library can be directly used for single cell whole transcriptome sequencing or targeted sequencing workflows