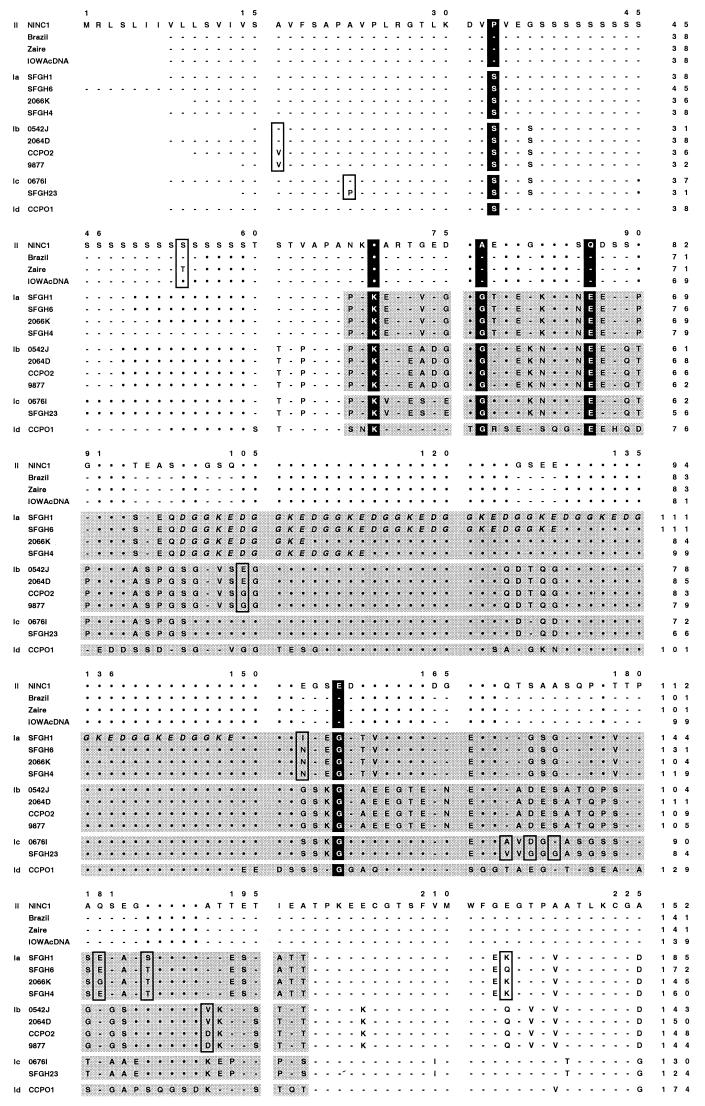
FIG. 8.
gp15/45/60 protein sequences are extremely polymorphic. The deduced gp15/45/60 amino acid sequences of 23 human and animal isolates were aligned using the ClustalW algorithm, and the sequence of the NINC1 animal isolate was used as a reference to which all others were aligned. The gp15/45/60 sequences from Iowa, NT009, KSU1, Peru, UCP, and TAMU were identical, and only the Iowa sequence is shown; likewise, only the first sequence from each of the identical pairs, 0542J and 0541L, SFGH4 and SFGH3, and 9877 and NEMC1, is presented. The five allelic groupings are explicitly indicated in the left margin by II, Ia, Ib, Ic, and Id. Hyphens in the alignment indicate identical residues, and filled circles indicate gaps; boxed residues depict SAAPs among sequences within a particular allelic grouping. The eight residues strictly conserved within either the allele I or allele II groupings are vertically highlighted throughout all sequences in black. The hypervariable region present within the gp45 sequence of human isolates (allelic groups Ia to Id) is indicated by shading. The tandemly repeated DGGKE sequence found within this region in all genotype Ia isolates is italicized. The two glutamic acid residues found in the putative protease cleavage site of the gp60 precursor are boxed throughout. The numbering above the sequences includes the gaps introduced for alignment purposes, while the numbers at the end of each line denote the amino acid position within that particular sequence.