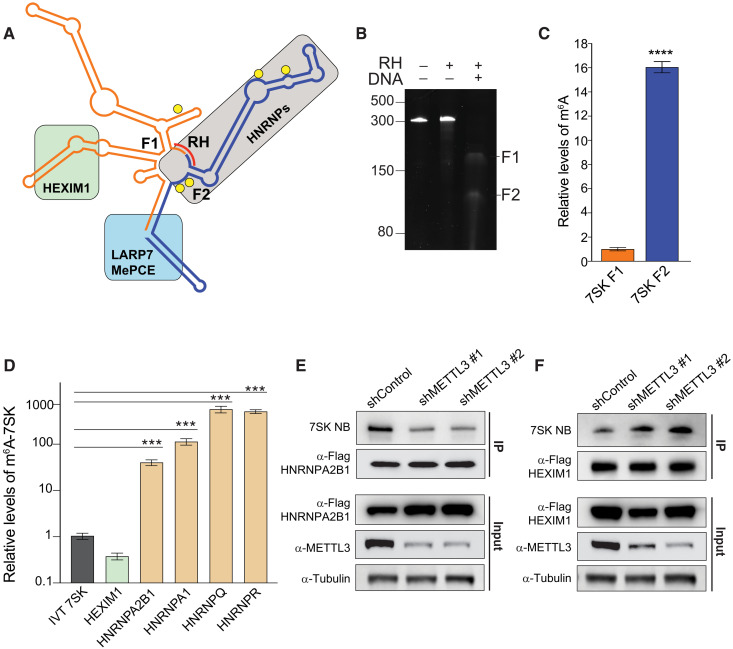
Fig. 2. HEXIM1 preferentially binds unmethylated 7SK.
(A) 7SK secondary structure representation. Proteins known to interact with 7SK are depicted as a green (HEXIM1/P-TEFb), gray (HNRNPs), or light blue (LARP7 and MePCE) box. Predicted m6A motifs are shown as yellow circles, and the complementary DNA oligo used to fragment 7SK in the RNase H (RH) digestion is shown as a red line. (B) RNase H digestion of endogenous 7SK. Tris-borate EDTA (TBE)–urea acrylamide RNA gel showing untreated 7SK and treated with RNase H, with or without the addition of the complementary DNA oligo (DNA). (C) Relative m6A levels in the two fragments generated by RNase H treatment of endogenous 7SK pulled down using Flag-tagged LARP7 as shown in (B). After purification, 7SK was digested by RNase H using the DNA complementary oligo depicted in (A). After fragmentation, the purified fragments were subjected to m6A-IP followed by qRT-PCR quantification. The m6A-immunoprecipitated 7SK fragments were normalized to each fragment before the m6A-IP. The graph shows means ± SD from three biological replicates. Student’s two-tailed t test; ****P < 1 × 10−4 (D) Stably expressing Flag-tagged proteins were used to pull down endogenous 7SK. After purification of the pulled-down 7SK RNA, a second IP using an m6A antibody was performed, and the levels of 7SK were quantified by qRT-PCR. Nonmethylated IVT 7SK RNA was used as a negative control for the m6A-IP. The graph shows means ± SD from three biological replicates. One-way ANOVA with Dunnett’s posttest; ***P < 1 × 10−3. (E and F) 7SK-protein interaction with HNRNPA2B1 (E) and HEXIM1 (F) upon METTL3 depletion using two independent shRNAs. The top two panels represent the pull-down of endogenous 7SK measured by Northern blot. The immunoprecipitated proteins are detected by Western blot using the indicated antibodies. The bottom three panels show the input protein levels before the IP. Blots are representatives of at least two biological repeats.