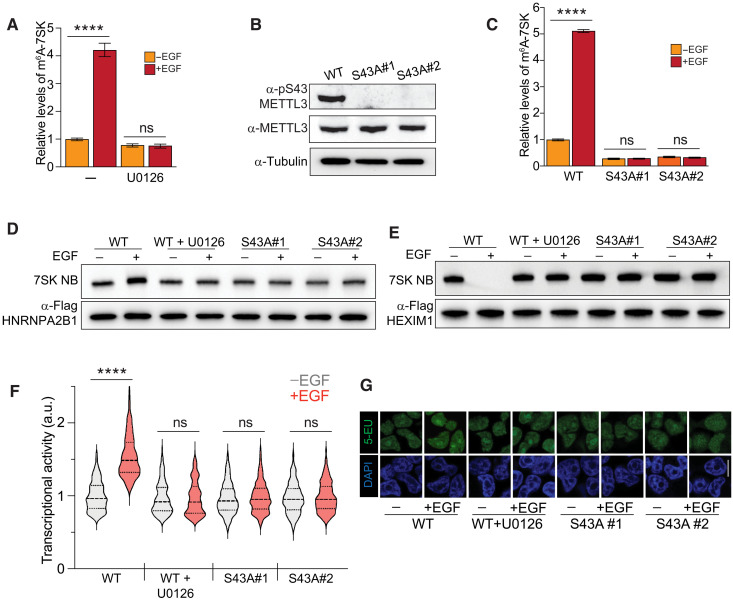
Fig. 5. EGF transcriptional activation depends on METTL3 phosphorylation.
(A) Effect of U0126 on the EGF-induced m6A methylation of 7SK. The experimental design is similar to Fig. 4A. m6A-methylated RNA was immunoprecipitated from cells under the treatments shown in the figure, and 7SK was quantified by qRT-PCR. The m6A-immunoprecipitated 7SK RNA was normalized to the total 7SK. The graph shows means ± SD from three biological replicates. Two-way ANOVA with Tukey’s posttest; ****P < 1 × 10−4. (B) CRISPR-Cas9 mutagenesis of S43A of METTL3. Western blot of METTL3 pS43 upon EGF stimulation in wild-type (WT) cells and two METTL3S43A homozygous clones. (C) Similar to (A), but in this case, wild-type cells and two METTL3S43A homozygous clones were used. The graph shows means ± SD from three biological replicates. Two-way ANOVA with Tukey’s posttest; ****P < 1 × 10−4. (D and E) 7SK-protein interaction with HNRNPA2B1 (D) and HEXIM1 (E), upon EGF stimulation in wild-type cells and two independent METTL3S43A homozygous clones. Blots are representatives of two biological replicates. (F and G) Effect of U0126 and mutagenesis of S43A-METTL3 on transcriptional activity induced by EGF stimulation, as measured by RNA Click-iT. Violin plots of fluorescent measurement of 600 cells per condition. (G) Micrographs of representative cell nuclei of (F). Scale bar, 10 μm. Two-way ANOVA with Tukey’s posttest; ****P < 1 × 10−4.