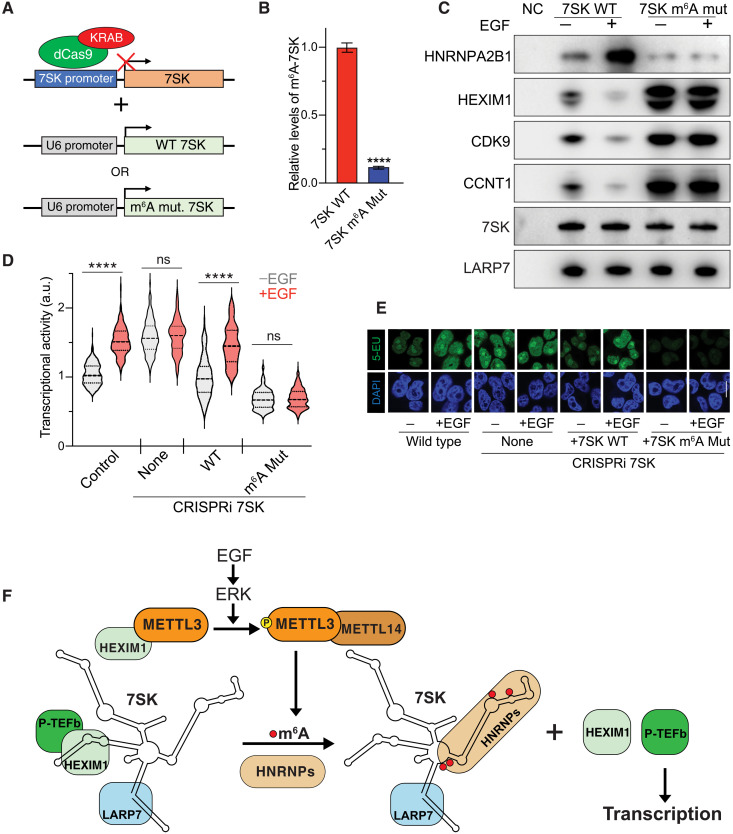
Fig. 6. 7SK m6A methylation is required for HEXIM1 dissociation and transcriptional activation downstream EGF signaling.
(A) CRISPRi was used to down-regulate the expression of 7SK. Once cells were depleted of endogenous 7SK, lentiviral vectors were used to stably express exogenous wild-type or m6A-mutant 7SK under the U6 promoter. (B) IP of m6A-methylated RNA followed by qRT-PCR of 7SK from cells depleted of 7SK and transduced with wild-type 7SK (red bar) or 7SK m6A mutant (blue bar). The m6A-immunoprecipitated 7SK RNA was normalized to total 7SK. The graph shows means ± SD from three biological replicates. Student’s two-tailed t test; ****P < 1 × 10−4 (C) Flag-LARP7 IP of CRISPRi 7SK plus wild-type 7SK or 7SK mutant for m6A sites upon EGF stimulation. Western blot for the interacting proteins using the indicated antibodies. Northern blot using a probe against 7SK was performed. (D and E) Effect of mutagenesis of m6A sites of 7SK on transcriptional activity induced by EGF stimulation, as measured by RNA Click-iT. Violin plots of fluorescent measurement of 600 cells per condition. (E) Micrographs of representative cell nuclei of (D). Scale bar, 10 μm. Two-way ANOVA with Tukey’s posttest; ****P < 1 × 10–4. (F) Model. The work presented here is summarized in this model. Upon EGF stimulation, the downstream effector ERK kinase is activated and phosphorylates METTL3 at position S43. This phosphorylation results in the release of METTL3 from its association with HEXIM1, thus allowing the methylation of 7SK. Subsequently, m6A methylation of 7SK results in its association with HNRNP proteins and release of the HEXIM1/P-TEFb complex. Free P-TEFb is then available to induce transcription activation.