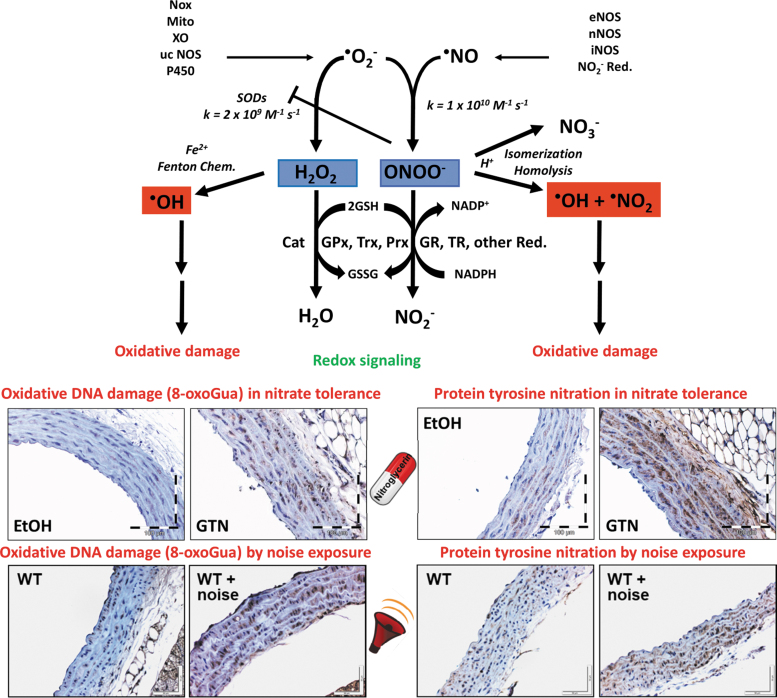
FIG. 1.
The chemical basis of vascular oxidative stress and redox signaling by superoxide and nitric oxide in animal models of nitrate tolerance and traffic noise exposure. Upper part: Superoxide (•O2−) formation from Nox, the mitochondrial respiratory chain (Mito), XO, a ucNOS, and P450 side reactions confer redox signaling mainly upon breakdown by self-dismutation or catalyzed by SODs to hydrogen peroxide. Reaction with thiol groups is a major route of detoxification for H2O2 via Prx and Trx or the selenol in GPx—these systems require energy-consuming recycling by NADPH-coupled reductases (GR, TR). Another route for H2O2 decomposition is catalyzed by catalase (Cat). When H2O2 accumulates, it may lead to the Fenton reaction (hydroxyl radical formation) upon reaction with ferrous iron (Fe2+), triggering severe oxidative damage at the protein, lipid, and DNA levels. Likewise, nitric oxide (•NO, previously termed EDRF) is formed from eNOS, nNOS, iNOS, or reduction of nitrite from nutrition conferring the diffusion-controlled reaction with superoxide to yield peroxynitrite anion (ONOO−). This rapid kinetics even outcompetes the extremely fast SOD-catalyzed breakdown of superoxide. Upon protonation, peroxynitrite undergoes either spontaneous isomerization to nitrate (representing a detoxifying route) or undergoes homolysis to form the hydroxyl radical (•OH) and the nitrogen dioxide (•NO2) radical leading to oxidative damage comparable with the Fenton reactivity of H2O2. Lower part: (Left) Hydroxyl radicals typically induce oxidative DNA damage in the form of 8-oxoGua as envisaged in GTN-treated, nitrate-tolerant rats (GTN with a dose of 50 mg/kg/day for 3.5 days) or aircraft noise-exposed mice [72 dB(A) around-the-clock for 4 days] by immunohistochemistry (brown staining). (Right) ONOOH and derived free radicals typically induce protein tyrosine nitration as envisaged in GTN-treated, nitrate-tolerant rats or noise-exposed mice by immunohistochemistry (brown staining), although MPO/H2O2/nitrite reaction will also lead to nitration. Scheme in the upper part summarized from Daiber and Ullrich (2002) and adapted from Daiber et al (2020c) and Daiber et al (2014) with permission. Images in the lower part were reproduced from Mikhed et al (2016) (for nitrate tolerance) and Kvandova et al (2020) (for noise exposure) with permission. 8-oxoGua, 8-oxoguanine; EDRF, endothelium-derived relaxing factor; eNOS, endothelial nitric oxide synthase; GPx, glutathione peroxidase; GR, glutathione reductase; GTN, nitroglycerin; iNOS, inducible nitric oxide synthase; MPO, myeloperoxidase; nNOS, neuronal nitric oxide synthase; Nox, NADPH oxidases; Prx, peroxiredoxins; SODs, superoxide dismutases; TR, thioredoxin reductase; Trx, thioredoxin; ucNOS, uncoupled nitric oxide synthase; XO, xanthine oxidase.