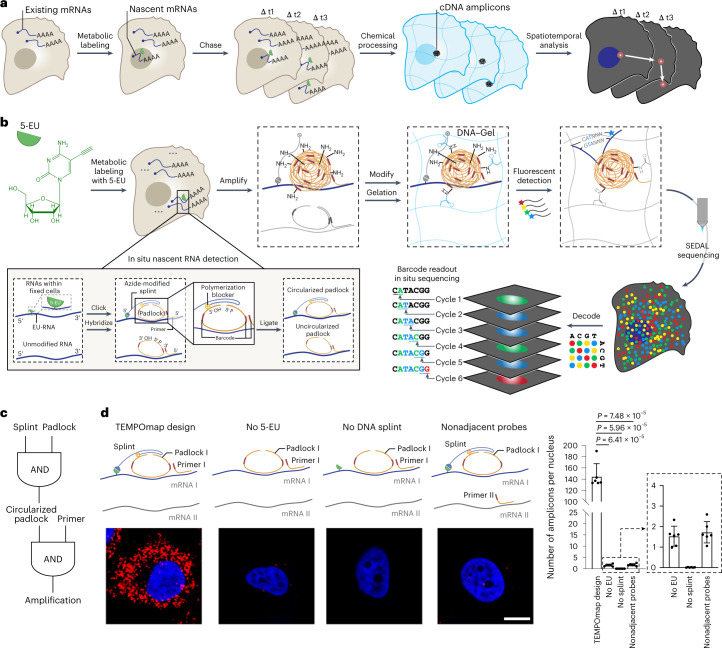
Fig. 1. TEMPOmap enables spatiotemporally resolved transcriptomics.
a, Overview of TEMPOmap pipeline: nascent RNAs of several timepoints are collected and in situ sequenced, followed by spatiotemporal RNA analyses. b, TEMPOmap experimental workflow. After 5-EU-labeled cells are prepared, a set of tri-probes (splint, primer and padlock) are conjugated or hybridized to cellular mRNAs (Extended Data Fig. 1b,i for more details), resulting in the enzymatic replication of each padlock sequence into cDNA amplicons. The amplicons are anchored in situ via a functionalized acrylic group (blue) to a hydrogel mesh to create a DNA-gel hybrid (blue wavy lines). The five-base barcode on each amplicon is read out by six rounds of SEDAL. Thus, multiplexed RNA quantification reveals gene expression in nascent subcellular locations. c, DNA tri-probe design rationale. The generation of an amplicon requires the presence of splint, circularized padlock and primer probes in proximity. d, Left, schematics and representative fluorescent cell images of negative control experiments of c, showing three-part probe requirement for signal amplification. mRNA_I represents ACTB and mRNA_II represents GAPDH. All four images show ACTB (red) mRNA in HeLa cells (DAPI in blue). Right, quantification of cell images showing the average amplicon reads per cell (n = 6 images were measured containing 310, 585, 386 and 421 cells for each condition from left to right, respectively). Two-tailed t-test. Data shown as mean ± s.d. Scale bar, 10 µm.