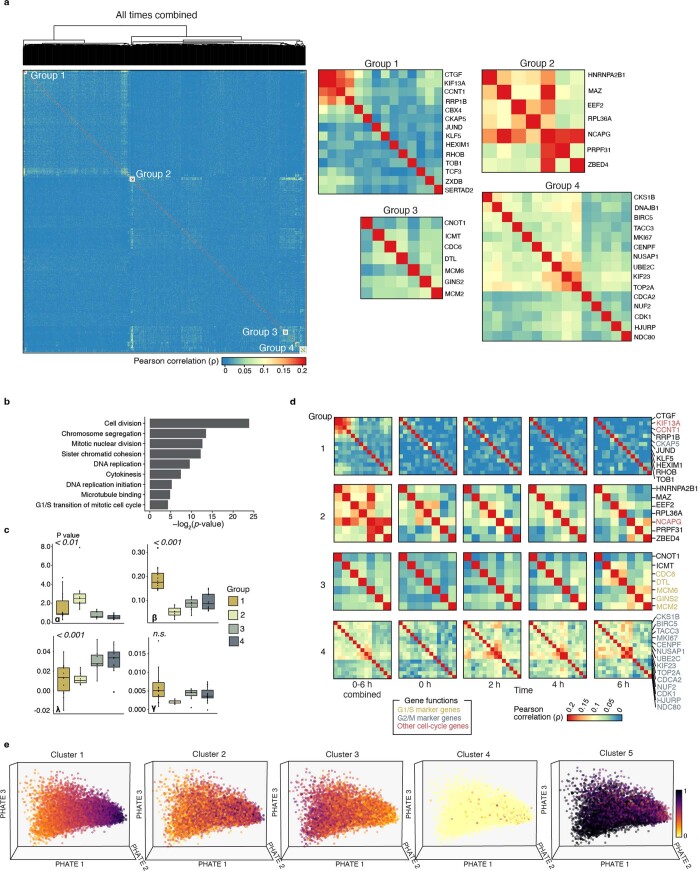
Extended Data Fig. 6. Single-cell RNA expression co-variation analysis.
a, Heatmap depicting the pairwise correlation of 911 genes by TEMPOmap-measured single-cell RNA covariation when combining four timepoints (0, 2, 4, 6 hrs chase), where the color indicates the value of Pearson correlation. Group 1-4 are highlighted for highly correlated gene modules (left) and zoomed in (right). b, Pathway enrichment analysis result of genes in Group 1-4 from single-cell gene covariation heatmap in a using DAVID. c, Boxplots showing the distribution of six kinetic parameters of four gene groups (n = 14 genes in Group 1; n = 7 genes in Group 2, n = 7 genes in Group 3, n = 15 genes in Group 4). Units of α is RNA concentration/hr, where RNA concentration is defined by copies/voxel (voxel = 200 nm x 200 nm x 350 nm). Units of β, λ, γ are hr−1. P values, one-way ANOVA test. Data shown as means, 25-75% quartiles and ranges. d, Heatmaps were generated showing matrices of the pairwise gene co-variation in each of the 0, 2, 4 and 6 hrs chase timepoints. Gene order along each matrix was the same and determined by the hierarchical clustering tree of the matrix combining the four timepoints in a (results were not shown). Zoom-in views of Group 1-4 from the co-variation heatmaps generated by gene expression in each timepoint, showing the correlation of RNA co-variation of each gene module across individual timepoints. Color in heatmaps indicates the value of Pearson correlation. G1/S and G2/M marker genes were annotated in each gene block. e, PHATE visualization of single cells showing the expression level (the fraction of reads in the cluster/total reads) of Cluster 1-5.