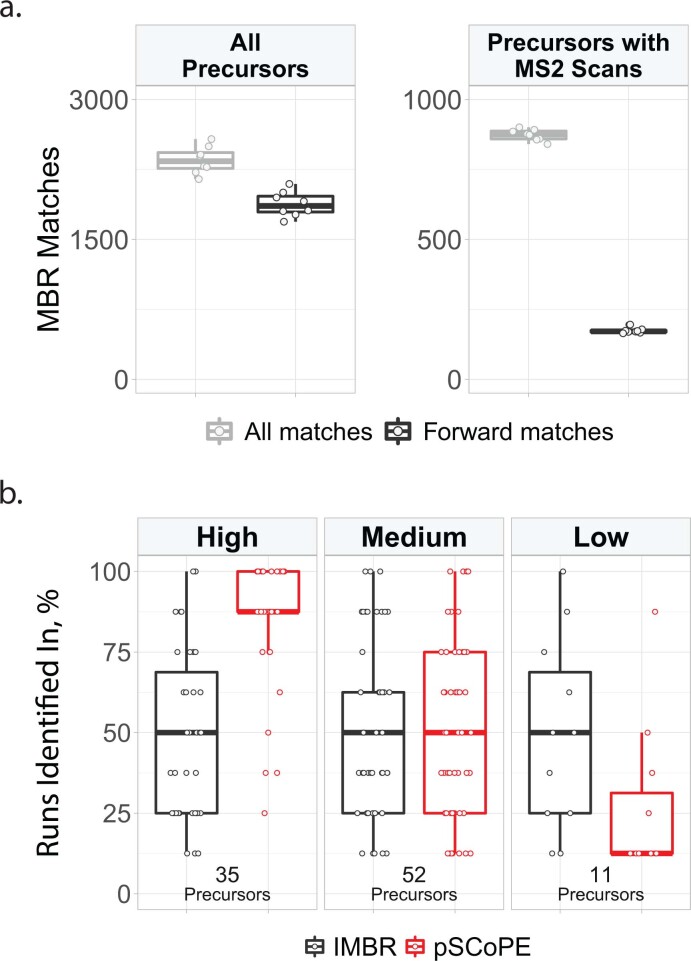
Extended Data Fig. 5. pSCoPE outperforms isobaric Match Between Runs (iMBR) for increasing consistency of identification across single-cell experiments.
(a) The ‘All Precursors’ facet heading indicates the total number of MBR-facilitated precursor identifications in each of 8 shotgun analyses. The ‘Precursors with MS2 Scans’ facet heading indicates the total number of MBR-facilitated precursor identifications that are associated with MS2 scans, enabling reporter ion quantitation. In both facets, the identifications are segmented into ‘All matches’, a category which includes matches to reverse sequences, and ‘Forward matches’, which does not. Each point represents an experiment. Data derived from shotgun experiments shown in Fig. 2a–e. (b) The intersected precursors between the MBR-facilitated forward sequence matches and the corresponding prioritized analyses were then compared based on consistency of identification across the 8 experiments associated with each acquisition method. Each point represents a precursor. Data derived from shotgun and pSCoPE analyses shown in Fig. 2a. Boxplot whiskers display the minimum and maximum values within 1.5 times the interquartile range of the 25th and 75th percentiles, respectively; the 25th percentile, median, and 75th percentile are also featured.