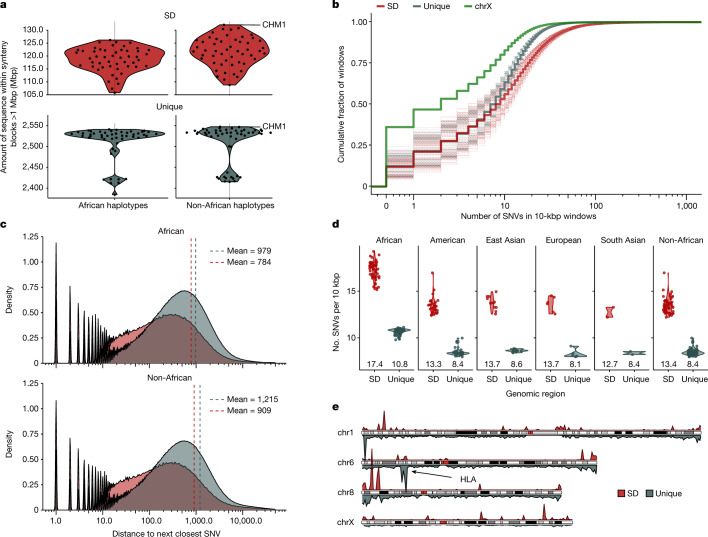
Fig. 1. Increased single-nucleotide variation in SDs.
a, The portion of the human genome analysed for SD (red) and unique (blue) regions among African and non-African genomes. Shown are the number of megabase pairs aligned in 1:1 syntenic blocks to T2T-CHM13 v1.1 for each assembled haplotype. Data are shown as both a single point per haplotype originating from a single individual and a smoothed violin plot to represent the population distribution. b, Empirical cumulative distribution showing the number of SNVs in 10-kbp windows in the syntenic regions stratified by unique (grey), SD (red) and the X chromosome (chrX; green). Dashed lines represent individual haplotypes and thick lines represent the average trend of all the data. c, Distribution of the average distance to the next closest SNV in SD (red) and unique (grey) space separating African (top) and non-African (bottom) samples. Dashed vertical lines are drawn at the mean of each distribution. d, Average number of SNVs per 10-kbp window in SD (red) versus unique (grey) space by superpopulation and with mean value shown underneath each violin. The non-African column represents an aggregation of the data from all non-African populations in this study. e, Density of SNVs in 10 bp of each other for SD (top, red) and unique (bottom, grey) regions for chromosomes 1, 6, 8 and X comparing the relative density of known (for example, HLA) and new hotspots of single-nucleotide variation.