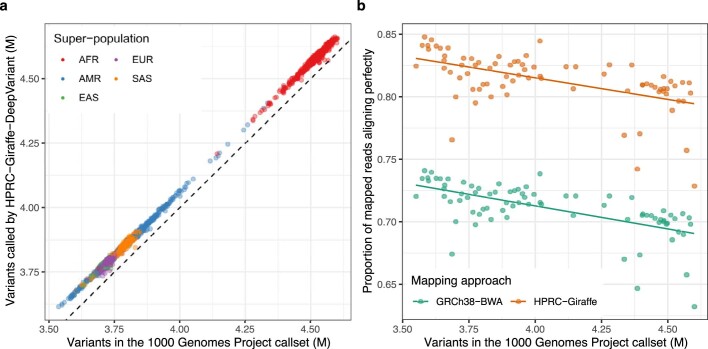
Extended Data Fig. 7. Performance comparison of pangenome-based variant calling and read mapping across populations.
a, Number of variants with at least one alternate allele (i.e. excluding homozygous for the reference allele) for each in the 1KG samples. The number of variants in the 1KG callset (x-axis) are compared to the variants found when aligning reads to the HPRC pangenome and calling variants with DeepVariant (y-axis). Points (samples) are coloured by their super-population label from the 1KG. b, The proportion of mapped reads that align perfectly (y-axis) is shown for a subset of samples from the 1KG, ordered by the number of variants called (x-axis). Two mapping approaches are compared: mapping short reads to GRCh38 with BWA (green); mapping to the HPRC pangenome with Giraffe (orange). The samples were selected to span the x-axis.