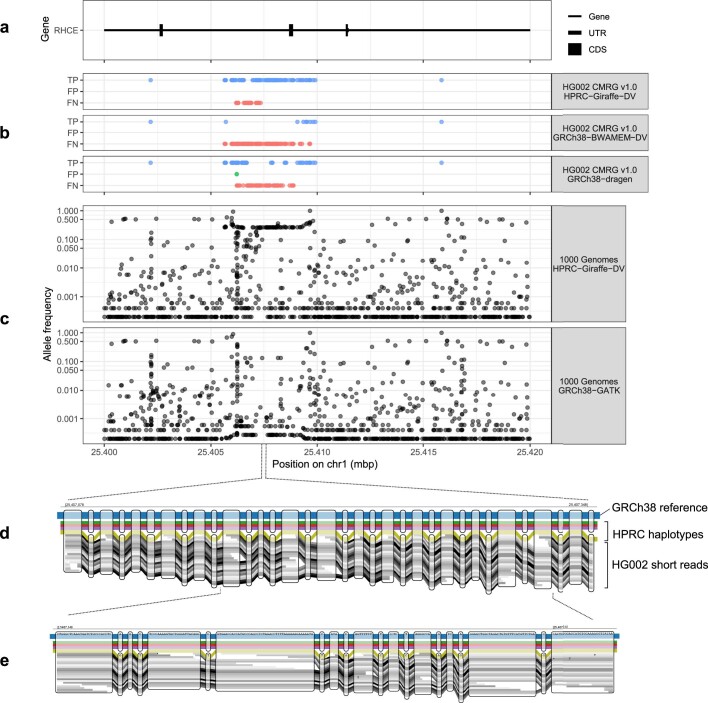
Extended Data Fig. 8. Improved genotyping in the challenging medically-relevant gene RHCE.
a, Gene annotation of part of the RHCE gene. b, Genotyping performance in this region for three approaches (horizontal panels). The top panel, using the HPRC pangenome, shows the best performance with most variants being true positives (TP, blue points) based on the CMRG (v.1.0) truth set while more other methods have a higher number of false negatives (FN, red points). c, Allele frequency across 2,504 unrelated individuals of the 1KG. The HPRC-Giraffe-DeepVariant calls show higher frequencies. In particular, the gene-converted alleles, at about 25.406-25.410 Mbp, are observed at ~25% frequency, similar to estimates from the HPRC haplotypes (Fig. 5a–c). d,e, A pangenomic view of the gene-converted region showing 1 of 4 haplotypes in the HPRC pangenome supporting the non-reference alleles. The inclusion of this haplotype in the HPRC pangenome enables short sequencing reads, here from HG002, to map along this gene-converted haplotype.