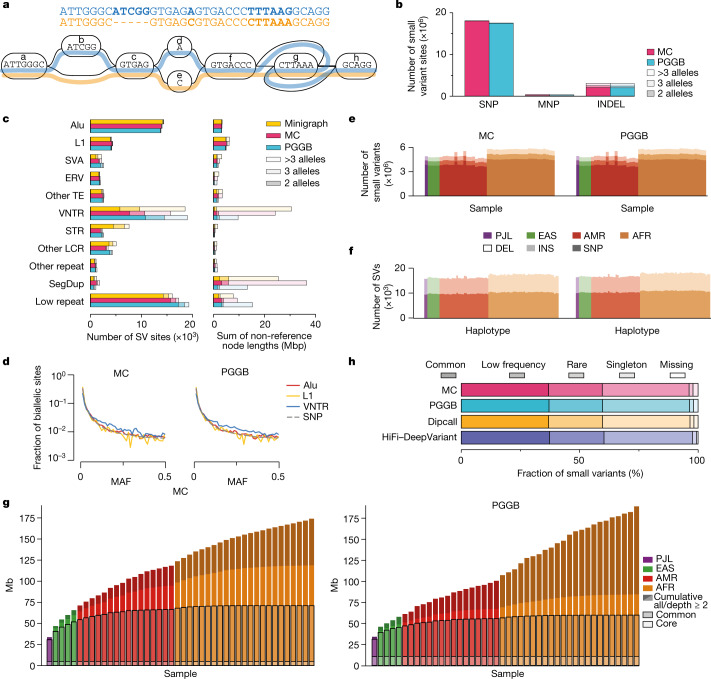
Fig. 3. Pangenome graphs represent diverse variation.
a, A pangenome variation graph comprising two elements: a sequence graph, the nodes of which represent oriented DNA strings and bidirected edges represent the connectivity relationships; and embedded haplotype paths (coloured lines) that represent the individual assemblies. b, Small variant sites in pangenome graphs stratified by the variant type and by the number of alleles at each site. MNP, multinucleotide polymorphism. c, SV sites in the pangenome graphs stratified by repeat class and by the number of alleles at each site. Other TE, a site involving mixed classes of transposable elements (TEs). VNTR, variable-number tandem repeat, a tandem repeat with the unit motif length ≥7 bp. STR, short tandem repeat, a tandem repeat with the unit motif length ≤6 bp. Other LCR, low-complexity regions with mixed VNTR and STR and low-complexity regions without a clear VNTR or STR pattern. Other repeat, a site involving mixed classes of repeats. SegDup, segmental duplication. Low repeat, a small fraction of the longest allele in a site involving repeats. d, Pangenome minor AF (MAF) spectrum for biallelic SNP, VNTR, L1 and Alu variants in the MC and PGGB graphs. e,f, Number of autosomal small variants per sample (e) and SVs per haplotype (f) in the pangenome. Variants were restricted to the Dipcall-confident regions. Samples are organized by 1KG populations. g, Pangenome growth curves for MC (left) and PGGB (right). Depth measures how often a segment is contained in any haplotype sequence, whereby core is present in ≥95% of haplotypes, common is ≥5%. h, Small variants in the GIAB (v.3.0) ‘easy’ regions annotated with AFs from gnomAD (v.3.1.2).