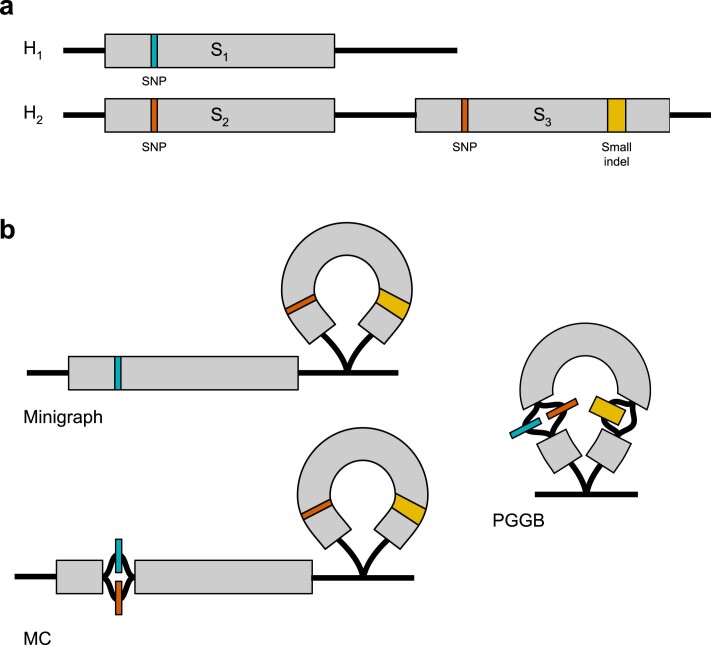
Extended Data Fig. 3. The differences in pangenome graph construction methods for Minigraph, MC, and PGGB.
a, Two haplotypes (H1 and H2) vary in copy number of a chromosomal segment S. The S1, S2, and S3 segments are highly similar with only a SNP or a small indel. b, Pangenome graph structures for Minigraph, MC, and PGGB. Minigraph used H1 as an initial backbone and then augmented with SVs (≥50 bp) from H2, such that the SNP in S2 is not represented in the pangenome graph. MC added small variants (<50 bp) to the pangenome graph constructed by Minigraph. PGGB used a symmetric, all-by-all alignment of haplotypes to build a pangenome graph whose structure is not affected by the order of inputs (unlike Minigraph and MC). The critical difference in graph construction is that, due to ambiguous pairwise relationships of paralogs, PGGB tends to collapse copy-number polymorphic loci like segmental duplications and VNTRs into a single copy through which haplotypes loop, while Minigraph and MC do not.