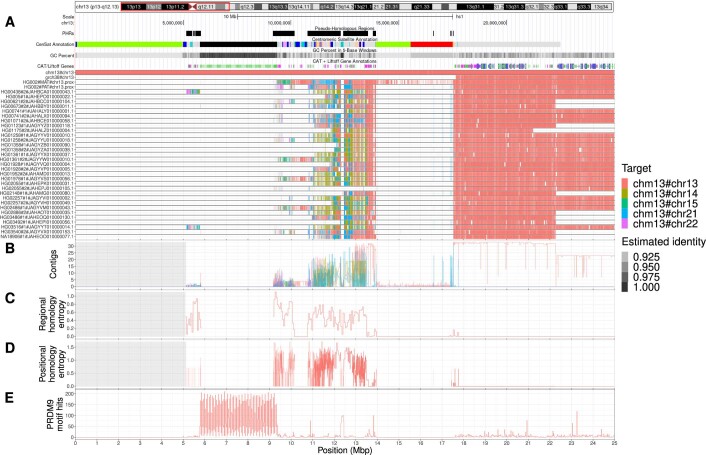
Extended Data Fig. 4. Characteristics of the pseudo-homologous regions of acrocentric chromosomes on chromosome 13.
(A) We focus on the first 25 Mbp of chromosome 13, shown here as a red box over T2T-CHM13 cytobands. Pseudo-homologous regions (PHRs), where diverse sets of acrocentric chromosomes recombine, are highlighted relative to T2T-CHM13 genome annotations for repeats, GC percentage, and genes. Above, we indicate regions of interest described in the main text: rDNA, SST1 array, centromere, and q-arm. Below, we show T2T-CHM13-relative homology mosaics for each chromosome 13 matched contig from HPRCy1-acro, with the most-similar reference chromosome at each region shown using the given colors (Target). (B) Aggregated untangle results in the SAACs. For each acrocentric chromosome, we show the count of its HPRCy1 q-arm-anchored contigs mapping itself and all other acrocentrics (Contigs), (C) as well as the regional (50kbp) untangle entropy metric (Regional homology entropy) computed over the contigs’ T2T-CHM13-relative untanglings. (D) By considering the multiple untangling of each HPRCy1-acro contig, we develop a point-wise metric that captures diversity in T2T-CHM13-relative homology patterns (Positional homology entropy), leading to our definition of the PHRs. (E) The patterns of homology mosaicism suggest ongoing recombination exchange in the SAACs. A scan over T2T-CHM13 reveals that the rDNA units are enriched for PRDM9 binding motifs, and thus may host frequent double stranded breaks during meiosis. In (B-D) a gray background indicates regions with missing data due to the lack of non-T2T-CHM13 contigs. We provide the Centromeric Satellite Annotation (CenSat Annotation) track legend in Extended Data Table 1.