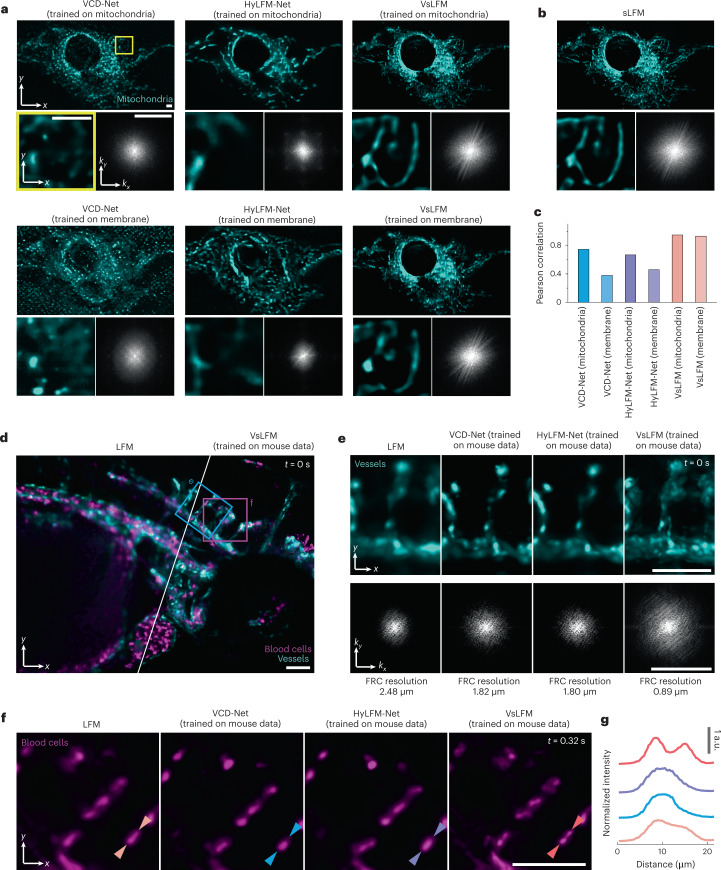
Fig. 4. VsLFM has better generalization ability than end-to-end networks.
a, MIPs and enlarged views of a fixed L929 cell with mitochondria labeling (TOM20-GFP), obtained by VCD-Net, HyLFM-Net and VsLFM trained on the same type of sample (mitochondria, upper row) and a different type of sample (membrane, lower row). The corresponding Fourier spectrum is shown in the bottom-right corner of each panel. b, Corresponding sLFM results as ground truth. c, Bar chart of Pearson correlations between the results of the mitochondria channel obtained by sLFM and the results obtained by VCD-Net, HyLFM-Net and VsLFM trained on different datasets. d, MIPs acquired by LFM (left) and VsLFM (right) of circulating blood cells (magenta) and vessels (cyan) in a zebrafish larva. The data were captured with a ×20/0.5 NA air objective at 50 vps. Given that the ground truth data cannot be obtained in this highly dynamic sample, the network models were trained on mouse liver data with vessel and neutrophil labeling and captured by a ×63/1.4 NA oil-immersion objective. e, Enlarged MIPs of the vessel channel marked by the blue box in d at t = 0 s, acquired by different methods. The corresponding Fourier transforms of the MIPs with estimated resolutions by Fourier ring correlation (FRC) are shown in the bottom row to indicate the resolution enhancement by VsLFM. f, Enlarged MIPs of the blood cell channel marked by the magenta box in d at t = 0.32 s. g, Normalized intensity profiles along the lines indicated by the arrows in f, showing that the two adjacent blood cells that could not be distinguished by previous methods were resolved by VsLFM. Scale bars, 5 μm (top, bottom left, a) and 3 μm−1 (bottom right, a), 50 μm (d,f), 50 μm (top) and 1 μm−1 (bottom) (e).