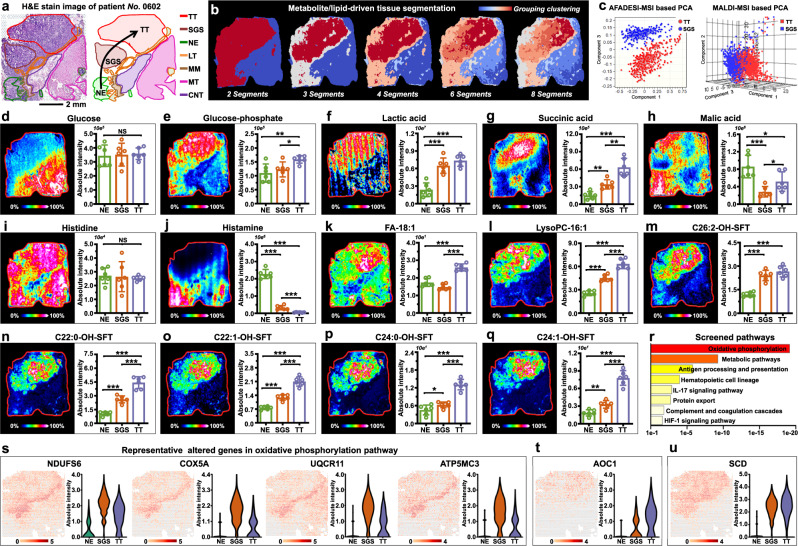
Fig. 5. Visualization of stepwise metabolic reprogramming in gastric cancer.
a H&E stain image of gastric cancer tissue section from patient “No.0602”, scale bar = 2 mm. The experiment was repeated three times. b Metabolite and lipid-driven tissue section segmentation. c PCA score plots based on AFADESI-MSI and MALDI-MSI data of tumor tissue (TT) and serrated glandular structure (SGS). d–q MS images and levels of glucose, glucose-phosphate, lactic acid, succinic acid, malic acid, histidine, histamine, FA-18:1, Lyso-PC-16:1, C26:2-OH-SFT, C22:0-OH-SFT, C22:1-OH-SFT, C24:0-OH-SFT, and C24:1-OH-SFT in different gastric cancer tissue section spots (seven tissue samples for spatial metabolomics and lipidomics, n = 6 independent section regions from patient “No.0602”, mean ± SD), ***p < 0.001, **p < 0.01, *p < 0.05, p-values were calculated using the unpaired two-tailed t test at confidence intervals 0.95, intensity in colour scale is relative value. r Pathways enriched in SGS tissue. s Representative altered genes in oxidative phosphorylation pathway, intensity in colour scale is log2 transformed. t, u Spatial expression images of AOC1 and SCD, intensity in colour scale is log2 transformed. TT Tumor tissue, SGS Serrated glandular structure, NE Normal epithelium, LT Lymphoid tissue, MM Muscularis mucosa, MT Muscle tissue, CNT Connective tissue.