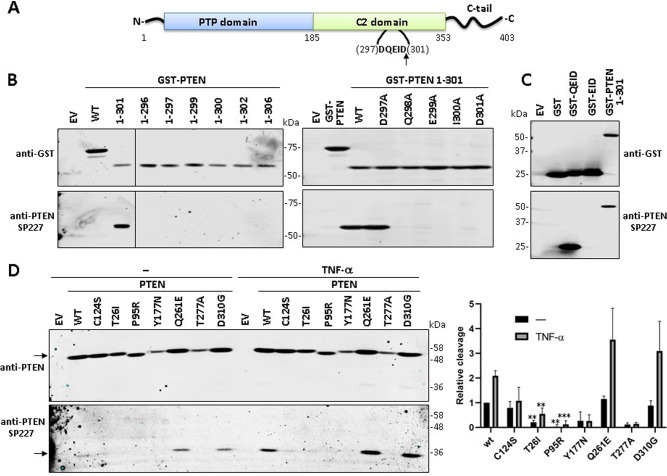
Fig. 5. Cleavage of PTEN PHTS variants at Asp301 as determined using the anti-PTEN SP227 mAb.
A Schematic of PTEN showing the caspase-3 cleavage motif (amino acids 297-301) and the Asp301 (D301) cleavage site at the unstructured loop at PTEN C2 domain. The arrow indicates the cleavage site. B Reactivity of the anti-PTEN SP227 mAb with GST-PTEN (N-terminal tagging) truncations. COS-7 cells were transfected with pRK5 plasmids encoding the indicated GST-PTEN truncations (EV empty vector, WT wild type), and cell lysates were analyzed by immunoblot using anti-PTEN SP227 or anti-GST (as a control of expression). In the right panel, the indicated amino acid substitutions, targeting the caspase-3 cleavage motif, were analysed in the background of GST-PTEN 1-301. C Reactivity of the anti-PTEN SP227 mAb with GST containing amino acids from the caspase-3 cleavage motif at PTEN C2 domain. COS-7 cells were transfected with pRK5 plasmids encoding the indicated GST fusion proteins (EV, empty vector), and cell lysates were analyzed as in B. D Cleavage of PTEN variants at Asp301. COS-7 cells were transfected with pRK5 plasmids encoding the indicated PTEN variants (EV empty vector, WT wild type), and then were kept untreated or were treated with TNF-α (50 ng/ml) for 6 h, followed by immunoblot using anti-PTEN SP227 or anti-PTEN 6H2.1 antibodies. The left panel shows a representative experiment (arrows indicate migration of PTEN [left upper panel] or PTEN 1-301 [left bottom panel]). The right panel shows the relative detection of PTEN bands from the PTEN variants as compared with PTEN wild type (WT) under untreated conditions (PTEN full-length and PTEN 1-301 normalized to 1), after quantification of the bands from three independent experiments. Results are shown as the ratio of the PTEN 1-301/PTEN bands (note that, for normalization, this ratio is = 1 in PTEN WT under untreated conditions). Statistical significance (Student’s t test P values) of the difference of some variants with respect to wild type is indicated with asterisks: ***p < 0.001, **p < 0.005.