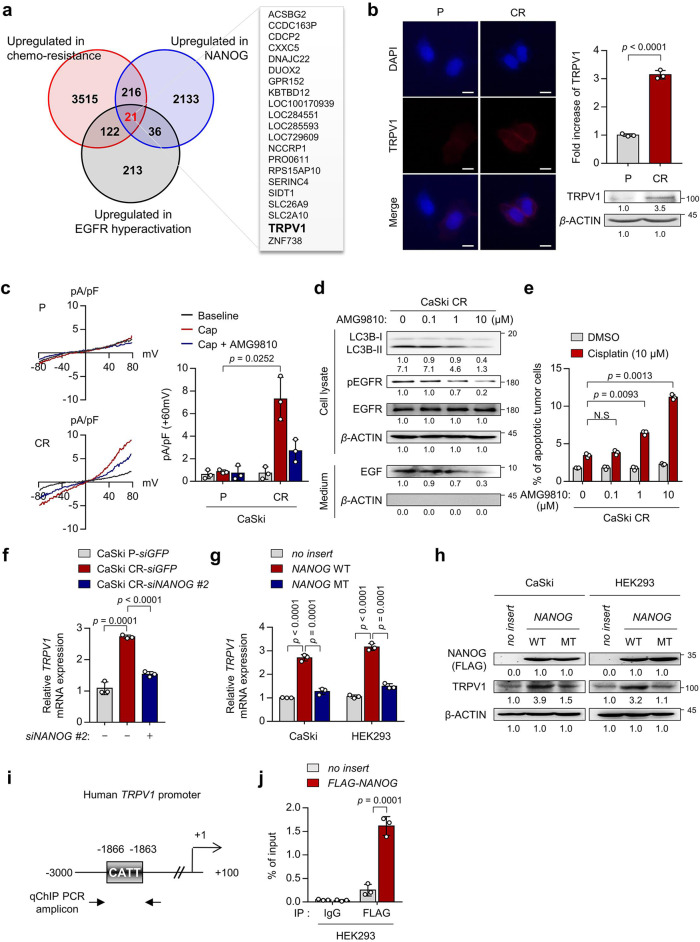
Fig. 4. NANOG directly regulates TRPV1 through promoter occupancy.
a Venn diagram showing the overlap of genes upregulated in high versus low chemotherapy-resistant cervical cancer patients (red), CaSki NANOG versus CaSki no insert (blue), and cervical cancer patients from the TCGA cohort classified as having high or low EGFR activity score (dark). b The cells were stained with anti-TRPV1 (red) antibodies and then visualized by confocal microscopy. DAPI was used to stain the nuclei. Scale bar, 10 μm. The graph depicts the experimental quantitation of TRPV1. The protein levels of TRPV1 and β-actin were confirmed by western blots. c Current-voltage relationships from capsaicin (Cap, red line) or AMG9810 (AMG, blue line)-induced whole-cell current in CaSki P versus CR cells. Summary of average outward current at +60 mV during Cap exposure in CaSki P versus CR cells. d The protein levels of secreted EGF and internal LC3B, pEGFR, and EGFR were confirmed by western blots. e Flow cytometry analysis of the frequency of apoptotic (active caspase 3+) cells after incubation with or without cisplatin for 24 h. f mRNA expression of TRPV1 was analyzed by qRT-PCR. g TRPV1 mRNA levels were measured by qRT-PCR. h Levels of TRPV1 and FLAG-NANOG proteins were confirmed by western blot. i Diagram of the human TRPV1 promoter region containing the NANOG-binding site. The arrows indicate the qChIP-PCR amplicon. j The cross-linked chromatin from HEK293 cells transfected with empty vector or FLAG-NANOG was immunoprecipitated with rabbit IgG or anti-FLAG antibodies. Relative enrichment of FLAG-NANOG in the TRPV1 promoter region was assessed by qChIP-PCR analysis with primers that amplified the genomic region indicated above. The ChIP data values represent the relative ratio to the input. Numbers below the blot images indicate the expression as measured by fold-change. All experiments were performed in triplicate. The p values by two-tailed Student’s t test (b), two-way ANOVA (c, e), or one-way ANOVA (f, g, j) are indicated. The data represent the mean ± SD. Source data are provided as a Source Data file. (NS, not significant).