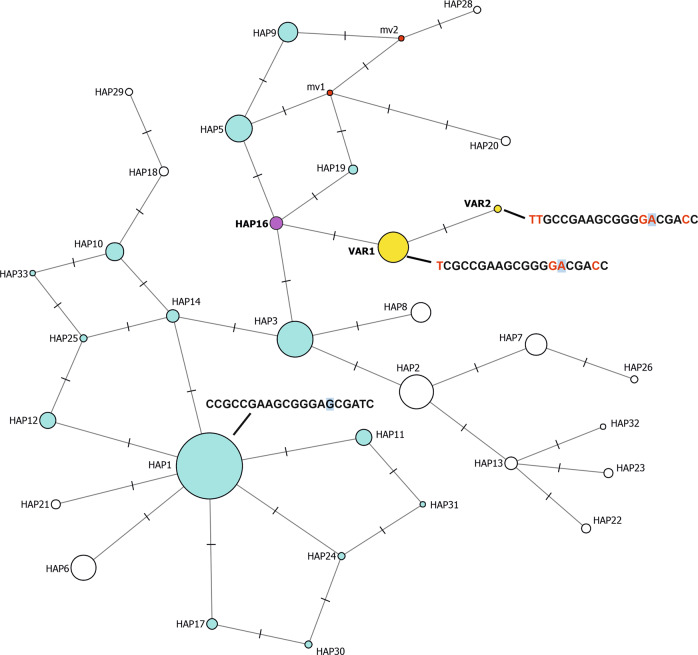
Fig. 4. Median-joining haplotype network constructed on a total of 136 individuals (N = 107 TSI and N = 29 comprising the maximum unrelated subset extracted from the study subjects), obtained including variants at all frequencies for finer dissection (including p.Val142Ile).
Ancestral haplotypes (forming the network torso) are represented in light blue (or purple). Ancestral haplotypes not detected in the analyzed sample set, but of which the existence is hypothesized are represented as “median vectors” (mv), in red. The two haplotypes with the p.Val142Ile variant are represented in yellow (VAR1 and VAR2) and the unique ancestral haplotype on which p.Val142Ile appeared in Tuscany is represented in purple (HAP16). Circle areas are proportional to the number of chromosomes carrying the haplotype, with HAP16 detected on 3 chromosomes in the TSI set and HAP1 (the most prevalent haplotype in TSI) found on 93. The full haplotype sequence is displayed for the most prevalent ancestral haplotype (HAP1) and for the two haplotypes with the p.Val142Ile variant. In the former, alleles that are different from those defining HAP1 are highlighted in red. The p.Val142Ile allele is displayed with a light blue background. Ticks along the network edges represent the number of differing alleles between the connected haplotypes. The full sequence of all haplotypes is provided in Supplementary Table 2 and details of all the haplotype-defining variants are reported in Supplementary Table 3.