Abstract
Genome-wide association studies (GWAS) conducted in European ancestry (EA) have identified hundreds of single-nucleotide polymorphisms (SNPs) associated with general cognitive function and/or Alzheimer’s disease (AD). The association between these SNPs and cognitive function has not been fully evaluated in populations with complex genetic substructure such as South Asians. This study investigated whether SNPs identified in EA GWAS, either individually or as polygenic risk scores (PRSs), were associated with general cognitive function and 5 broad cognitive domains in 932 South Asians from the Diagnostic Assessment of Dementia for the Longitudinal Aging Study in India (LASI-DAD). We found that SNPs identified from AD GWAS were more strongly associated with cognitive function in LASI-DAD than those from a GWAS of general cognitive function. PRSs for general cognitive function and AD explained up to 1.1% of the variability in LASI-DAD cognitive domain scores. Our study represents an important stepping stone toward better characterization of the genetic architecture of cognitive aging in the Indian/South Asian population and highlights the need for further research that may lead to the identification of new variants unique to this population.
Keywords: Dementia, Genetic risk score, India, Population structure, Single-nucleotide polymorphism
General cognitive function, a global measure of neurocognitive ability, tends to be relatively consistent throughout the life course and is a marker of healthy brain aging. Deficits in neurocognitive function represent an important component of neurodevelopmental and neuropsychiatric disorders, with neurocognitive impairments being the hallmark of conditions such as Alzheimer’s disease (AD). Genetic studies point to a strong heritable component of cognitive function and AD, with estimates being as high as 50%–80% (1,2). Identifying genetic variants that influence both cognitive function and neurocognitive disorders is critical for predicting the long-term risk of developing AD and other dementias and can shed light on underlying etiological pathways.
Genome-wide association studies (GWAS) conducted in participants of European ancestry (EA) have identified at least 161 AD-associated single-nucleotide polymorphisms (SNPs) from 90 loci in addition to APOE, which is the strongest genetic risk factor for AD in many populations (3–7). GWAS of AD conducted by Lambert et al. (3) and Kunkle et al. (5) identified 19 and 25 AD risk loci, respectively. A GWAS of clinically diagnosed AD and AD-by-proxy (71 880 cases, 383 378 controls) conducted by Jansen et al. (4) identified 29 risk loci, 12 of which were novel. Recently, 2 new GWAS of a similar outcome (AD/AD-by-proxy) conducted by Wightman et al. (6) and Bellenguez et al. (7) with sample sizes of 1 126 563 and 788 989, respectively, further identified 7 and 42 novel AD-associated loci (38 and 75 total loci). Polygenic risk scores (PRSs) for AD are known to be associated with cognitive impairment and/or decline and to a lesser extent with cognition in EA (8). Fewer GWAS have examined SNPs associated with cognition. The largest GWAS of general cognitive function, conducted by Davies et al. (9) in over 300 000 EA participants identified 178 SNPs from 148 independent loci. Many of these loci had been previously associated with cognition, educational attainment, AD, and/or other health outcomes. The transferability of these findings to cohorts of a different ancestry with notably different linkage disequilibrium patterns, such as South Asians, has not been fully evaluated.
In 2010, it was estimated that 4 million Indians over age 60 have dementia, with prevalence estimates likely to increase steadily as the population ages and life expectancy lengthens (10). Thus, it is urgent to better understand the role of known genetic risk factors for cognition and dementia in this population, as well as identify the genetic risk factors that are specific to India/South Asians. Evaluating whether the known genetic risk factors are associated with cognitive function in older adults who may show early signs of dementia may be particularly important. Currently, there are no large-scale GWAS of cognition and/or dementia in Indian or South Asian populations, and many of the known genetic risk factors were identified in the GWAS of EA participants. In previous work, we showed that PRSs from 3 EA AD GWAS (3–5) are associated with memory scores in older South Asians from the Harmonized Diagnostic Assessment of Dementia for the Longitudinal Aging Study in India (LASI-DAD) (11). However, the effects of novel loci identified from the recent AD GWAS (6,7) have yet to be determined. Meanwhile, cognitive function in older populations is characterized by both baseline cognitive ability and cognitive impairment/decline caused by neurodegeneration (12,13). In addition, cognitive function impacted by neurodegeneration is not limited to memory. Thus, further studies that evaluate the effects of both AD and cognition-associated genetic risk factors on multiple cognitive domains are needed to describe the genetic architecture of cognitive aging in South Asians.
In this study, we investigated whether independent genome-wide significant SNPs from GWAS for AD (3–7) and general cognitive function, (9) either by themselves or aggregated as PRS(s), were associated with measures of cognition in 932 older South Asians from LASI-DAD. We also estimated the variation in the cognitive domain scores explained by age, sex, education, PRSs for AD, and general cognitive function, and/or APOE ε2 and ε4 genotypes individually and simultaneously.
Method
Study Sample
The Longitudinal Aging Study in India (LASI) is a nationally representative sample of over 72 000 Indian adults aged 45 and over. The Harmonized Cognitive Assessment Protocol, informational interviews, and blood draws were administered to roughly 4 000 LASI respondents aged 60 years or older as part of the LASI-DAD add-on study. To ensure a study sample with a broad distribution of cognitive ability, a 2-stage stratified random sampling approach was employed to draw an equal number of respondents with high and low risks of cognitive impairment based on cognitive tests administered during the main LASI examination and proxy respondents (14). The LASI-DAD sample used for this analysis was drawn from 7 states and union territories across India (Supplementary Figure 1).
Cognitive Measures
The cognitive measures evaluated were based on the recent work by Gross et al. that derived 5 broad cognitive domains (orientation, memory, executive functioning, language/fluency, and visuospatial function) and a generalized cognitive domain (general cognitive function) in LASI-DAD using factor analysis (15). These cognitive domain scores were derived using methods based on item response theory and were internally standardized to an N(0,1) distribution in the full sample of 4 096 LASI-DAD participants.
Genotyping, Imputation, and Principal Component Analysis
A total of 960 participants from LASI-DAD were genotyped using the Illumina Infinium Global Screening Array-24 (GSA-24) BeadChip, Version 2.0 (Illumina), and the 1000 Genomes Project worldwide (16) reference panel (Phase 3, Version 5) was used to impute genotypes. Details of the quality control and the imputation protocol have been described previously (11) and are also outlined in the Supplementary Methods. As reported previously (11), the average imputation accuracy in LASI-DAD is relatively high (R2 > 0.8) for variants with minor allele frequency (MAF) > 0.1 (Supplementary Figure 2). For SNPs/variants with MAF < 0.1, and especially for variants with MAF < 0.02, the average imputation accuracy decreases sharply.
Principal component analysis (PCA) was performed in SNPRelate (17) to remove outliers and select a set of 932 unrelated individuals for analysis (also see (11)). We then merged the data with 3 publicly available data sets including 300 individuals from the Simon’s Genome Project (SGDP) (18), 2 504 individuals from 11 populations from the 1000 Genomes Project19, and 1 163 individuals from the Genome Asia Project (19). We performed PCA to study the structure within LASI-DAD samples alone, as well as in relation to worldwide populations of West Eurasians, East Asians, and Andamanese Islanders. For association analyses, we adjusted for population structure using the first 10 genetic PCs constructed in LASI-DAD samples alone.
Candidate SNP Selection and Calculation of Polygenic Risk Scores
Candidate SNPs investigated in this study included all independent genome-wide significant SNPs from 5 AD GWAS and a GWAS of general cognitive function (3–7,9), except those with poor imputation quality (R2 < 0.8) in LASI-DAD (Supplementary Table 1). In total, we selected 161 risk SNPs from AD GWAS plus 2 SNPs from APOE (rs429358 and rs7412). Of these, 1 SNP was excluded because it was not available in the 1000 Genomes Project reference panel, and 46 SNPs were excluded due to poor imputation quality (R2 < 0.8). Similarly, among the 178 candidate SNPs identified from the general cognitive function GWAS (9), 48 SNPs were removed due to poor imputation quality (R2 < 0.8). The present study thus analyzed a total of 116 AD risk SNPs and 130 general cognitive function risk SNPs. The APOE ε2 and ε4 alleles are haplotypes formed by the combination of rs7412 and rs429358. Given that the ε1 allele is extremely rare in Indian populations and worldwide (MAF << 0.01) (20,21), we used rs7412 (T) and rs429358 (C) as proxies for ε2 and ε4. Chromosome and position numbers of SNPs are provided for genome build GRCh37.
Five PRSs for AD (PRSAD_Lambert, PRSAD_Jansen, PRSAD_Kunkle, PRSAD_Wightman, and PRSAD_Bellenguez) were constructed from 5 AD GWAS in EA participants, respectively (3–7). Each AD PRS was computed independently using all reported independent genome-wide significant SNPs in each GWAS as , with β 𝑖 being the effect size associated with the risk allele for SNP i, and 𝑥 ij being the dosage of the risk allele for SNP i in individual j, as described previously (11). Variants in the APOE region were excluded and treated as an independent signal. Each PRS was then standardized to an N(0,1) distribution.
The PRS for general cognitive function (PRSGenCog) was created similarly, using the independent genome-wide significant SNPs from the general cognitive function GWAS in EA (12). Specifically, PRSGenCog was calculated as , with β 𝑖 being the effect size associated with the cognitive function-increasing allele for SNPi, and being the dosage of the same allele for SNP in individual . Since only z scores and p values were reported in the original GWAS, the effect size of each SNP was calculated using , where z is the z score, N is the sample size from the released summary statistics, and MAF is the minor allele frequency reported in the GWAS. The PRSGenCog was standardized to N(0,1).
Estimation of Genome-Wide Ancestry Proportions
We estimated the genome-wide proportion of Ancestral North Indian (ANI) ancestry using f4ratio test as implemented in ADMIXTOOLS (22). We used the model of population relationships shown in Moorjani et al. (23). The f4ratio test computes the ratio of f(YRI, Basque, test, Onge)/f(YRI, Basque, Georgian, Onge) which measures the excess of West Eurasian ancestry in a test sample compared to the Onge population. We ran the analysis where test = a single individual in LASI-DAD. This quantity was summed over all sites and the standard errors were computed using the block jackknife method (block size of 5 cM).
Estimation of Individual Autozygosity
We used PLINK v1.07 (24) to estimate individual autozygosity across the genome in a combined data set of LASI-DAD individuals and 3 non-Indian reference populations (CEU, CHB, and YRI) from the 1000 Genomes Project. PLINK uses a sliding window approach to find regions of the genome of at least 1 MB in length containing 100 contiguous homozygous SNPs. We allowed 1 heterozygous and 5 missing calls per segment. Runs of homozygosity (ROH) segments were identified separately for each individual. We applied this method to compute the cumulative sum of the ROH per individual.
Statistical Analysis
Proportion of variance in cognitive domain scores explained by demographic factors and population genetics
We examined how much variability in the cognitive domain scores was explained by demographic and population genetic factors including age, sex, education, genetic PCs, proportion of ANI ancestry (%ANI), and ROH. Specifically, for each cognitive measure, we first evaluated models that included only age, sex, or education. We then added the first 10 genetic PCs, %ANI, or ROH to a model including only age, sex, and education. To evaluate whether %ANI and/or ROH explained any additional variance in cognitive domain scores after adjusting for population structure using genetic PCs, we next evaluated the change in R2 after adding %ANI or ROH to a model including age, sex, education, and 10 PCs.
Allele frequency differences for candidate SNPs
To assess whether the allele frequencies of the candidate SNPs were different between LASI-DAD and the EA samples from GWAS, we used a 1-sample Z test. We considered the SNP allele frequencies to be significantly different if p < .05.
Association between candidate SNPs, PRSs, and cognitive domain scores
We tested the association between each AD or general cognitive function risk SNP and each of the 6 cognitive domain scores separately. Model 1 adjusted for age, sex, and the first 10 genetic PCs, and Model 2 additionally adjusted for education. For each cognitive measure, the false discovery rate (FDR) was calculated separately for AD and general cognitive function risk SNPs. We considered an SNP significantly associated if FDR q < 0.10. We next assessed whether each PRS was associated with each cognitive measure separately using Models 1 and 2. We considered a PRS significantly associated if p < .05.
Proportion of variance in cognitive domain scores explained by candidate SNPs and PRSs
We next examined how much variability in the cognitive domain scores was explained by APOE ε2 and ε4, the five AD PRSs, and PRSGenCog individually or simultaneously. Specifically, for each cognitive measure, we considered the model with age, sex, and 10 PCs to be the base model. Next, each genetic risk factor was added to the base model individually or in combination with other risk factors. Fully adjusted models included the base model and all other risk factors (education, APOE ε2 and ε4, and the AD and cognitive function PRSs).
Results
Descriptive Statistics
A total of 932 participants with both genotype and cognitive measures were included in the analysis. Males comprised 44% of the sample and the mean age was 69.3 (SD = 7.3) years (Table 1). Most participants (71%) had an education level less than lower secondary, 25% had upper secondary or vocational training, and 4% had tertiary education. The correlation among the broad cognitive domain scores ranged from 0.46 (visuospatial function and language/fluency) to 0.73 (executive function and orientation; Supplementary Table 2). The correlation between general cognitive function and the 5 broad domains ranged from 0.72 (visuospatial function) to 0.95 (executive function).
Table 1.
Sample Characteristics for LASI-DAD Participants (N = 932)
| Characteristic | Mean or No. | SD or % |
|---|---|---|
| Male sex | 409 | 44% |
| Age (y) | 69.3 | 7.34 |
| Education | ||
| Less than lower secondary | 658 | 71% |
| Upper secondary and vocational training | 235 | 25% |
| Tertiary | 39 | 4% |
| General cognitive function score | 0.11 | 0.93 |
| Broad cognitive domain scores | ||
| Orientation | 0.02 | 0.80 |
| Executive function | 0.11 | 0.90 |
| Language/fluency | 0.19 | 0.77 |
| Memory | 0.10 | 1.04 |
| Visuospatial function | -0.05 | 0.86 |
Note: LASI-DAD = Diagnostic Assessment of Dementia for the Longitudinal Aging Study in India.
Population Structure in LASI-DAD
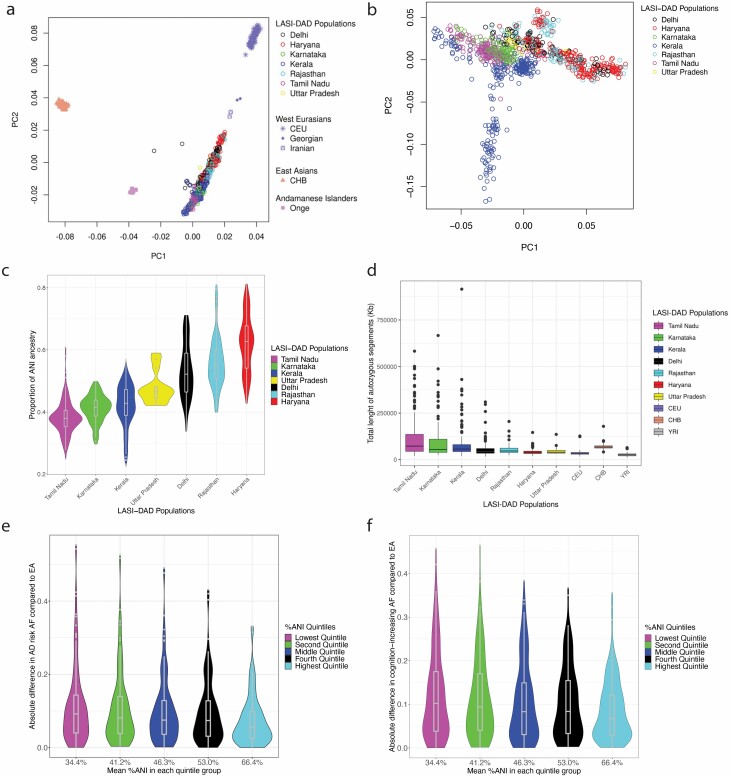
Recent genetic studies have shown that most South Asian groups descend from a mixture of 2 genetically divergent populations: ANI related to Central Asians, Middle Easterners, Caucasians, and Europeans; and Ancestral South Indians (ASI) not related to any groups outside the subcontinent and only distantly related to the indigenous Andamanese Islanders (23,25). To assess the population structure in LASI-DAD samples in relationship with other worldwide groups, we performed PCA with 932 individuals from 7 Indian states along with samples from 1000 Genomes Project including individuals of European (CEU) and East Asian (CHB) ancestry (16), as well as Andaman Islanders (Onge) from Genome Asia Project (19), Georgians and Iranians from SGDP. We observed that PC1 separated the West Eurasians (Georgians, Iranians, and Europeans) and East Asians (CHB), and the LASI-DAD individuals fall on a cline of relatedness to West Eurasians along PC2 (Figure 1A) (23,25). Our LASI-DAD sample, however, appears to be very heterogeneous in ancestry across regions or states, with Fst values ranging between 0.001 and 0.009. We also performed PCA with LASI-DAD samples alone and found that PC1 is correlated to the North/South axes in India, with the Northern regions of Delhi, Haryana, and Rajasthan being separated from the Southern states of Kerala and Tamil Nadu (Figure 1B, Supplementary Figure 3). PC2 primarily separated participants from Kerala into 2 clusters, which may represent distinct ancestry of some individuals currently living in Kerala and/or historical migration patterns. Within the limits of resolution, the 2 clusters from Kerala were not significantly different from each other in terms of demographic factors, %ANI ancestry, or ROH (all p > .05, see below for %ANI and ROH calculations). To quantify the proportion of ANI-ASI ancestry, we used the f4 ratio test using the population model described in Moorjani et al. (23). We inferred that ANI ancestry varies between ~38% and 62% across India (Figure 1C, Supplementary Table 3).
Figure 1.
Panel (A) shows the first 2 principal components (PCs) from the principal component analysis (PCA) of 932 LASI-DAD individuals from 7 Indian states or union territories with 3 reference populations including West Eurasians (Europeans [CEU], Georgians, Iranians), East Asians (Han Chinese), and Andamanese Islanders (Onge). Panel (B) shows the first 2 PCs from the PCA of 932 LASI-DAD individuals without reference populations. In both panels, PC1 is on the x-axis, and PC2 is on the y-axis for each participant. Color coding in panels (A) and (B) is according to the sampling location (state/union territory) of the participant. Panel (C) shows the Ancestral North Indian (ANI) ancestry in LASI-DAD populations. The f4ratio test was used to test the ANI ancestry proportion in LASI-DAD samples using ratio of f(YRI, Basque, test, Onge)/f(YRI, Basque, Georgian, Onge) where test = LASI-DAD samples belonging to each state/union territory in India. Standard errors were estimated using a block jackknife across genomes with 5-MB blocks across the genome. Panel (D) shows the genome-wide runs of homozygosity (ROH) in LASI-DAD. The cumulative sum of ROH per individual was estimated using PLINK for individuals from the 1000 Genomes Project (Africans [YRI], Europeans [CEU], and East Asians [CHB]) and 932 LASI-DAD samples from different states/union territories. Panels (E) and (F) show the distribution of the absolute difference in allele frequencies for 116 AD risk SNPs (E) and 130 general cognitive function SNPs (F) between quintile groups of ANI ancestry in LASI-DAD and European Ancestry (EA) GWAS samples. LASI-DAD participants were stratified into 5 groups based on quintiles of proportion of ANI (%ANI). The x-axis shows the mean %ANI of each stratified group in increasing order. Color coding is according to quintile. GWAS = genome-wide association study; LASI-DAD = Diagnostic Assessment of Dementia for the Longitudinal Aging Study in India
Previous studies have inferred strong founder events and considerable inbreeding in India (26,27). The inheritance of identical haplotypes from a common ancestor due to inbreeding or consanguinity can create long regions of homozygous genotypes known as ROH (28,29). To characterize the impact of inbreeding in our LASI-DAD samples, we estimated the total number of base pairs present in ROH across the genome using PLINK. We found South Asians have approximately one- to twofold excess of homozygous segments, compared to other worldwide populations (CEU, YRI, or CHB). On average, groups from South India (Tamil Nadu and Karnataka) had ~50% higher genome-wide ROH than Northern groups, though there was large variation within and across groups (Figure 1D).
Allele Frequency Distribution of Candidate SNPs
The allelic distributions of the 116 AD risk SNPs are presented in Supplementary Table 4. Briefly, most SNPs (98 out of 116) had a significantly different allele frequency between LASI-DAD and the EA GWAS samples (p < .05). However, although allele frequencies may be different across the 2 populations, the absolute difference is sometimes relatively small (e.g., <5%). Among the 98 AD risk SNPs with different allele frequencies, 65 SNPs (66.3%) were imputed, whereas a slightly lower proportion of SNPs with consistent allele frequencies were imputed (11 of 18 SNPs [61.1%]). Among the 130 cognition-associated SNPs, 103 had allele frequency differences between LASI-DAD and the EA GWAS sample (p < .05, Supplementary Table 5). Of the 103 SNPs, 89 (86.4%) were imputed. Of the remaining 27 SNPs with consistent allele frequencies, 26 (96.3%) were imputed.
To evaluate whether imputation to a primarily EA reference panel may have artificially reduced allele frequency differences, we tested whether the proportion of imputed SNPs differed between the SNPs with allele frequency differences compared to those without using Fisher’s exact test. The tests of proportion were not significant for the AD or the cognitive function risk SNPs. To examine whether allele frequency differences from EA were greater for the LASI-DAD samples that had a smaller %ANI, we calculated the absolute difference between the allele frequencies at each SNP for each %ANI quintile. The allele frequency differences tended to be smaller as the average proportion of ANI ancestry increased (Figure 1E and F).
Variability in cognitive domain scores explained by demographic factors and population genetics
We first examined how much variability in the cognitive domain scores was explained by demographic and population genetic factors including age, sex, education, genetic PCs, proportion of ANI ancestry (%ANI), and ROH as indicated by the change in R2 from corresponding regression models. The percentage of variance in the cognitive domain scores explained by age and sex alone ranged from 7.1% (visuospatial function) to 19.4% (orientation), and education added an additional 18.2% (orientation) to 34.7% (general cognitive function) (Supplementary Table 6). Beyond age, sex, and education, the additional variance explained by the first 10 genetic PCs ranged from 0.8% (visuospatial function) to 14.2% (language/fluency) with p < .05 for most cognitive domains except visuospatial function. The additional variance beyond age, sex, and education explained by %ANI and ROH was much lower (<0.001%–7.1%, and <0.001%–1.14%, respectively). However, %ANI was significant (p < .05) for 4 of the cognitive domain scores (general cognitive function, executive function, language/fluency, and memory) and ROH was significant for language/fluency. After controlling for age, sex, education, and the first 10 genetic PCs, %ANI and ROH do not explain any additional variance at p < .05, and thus were not included as covariates in association analyses.
Association Between Single SNPs and Cognitive Domain Scores
We assessed the association between each candidate SNP and cognitive measure in LASI-DAD using 2 models. Model 1 adjusted for age, sex, and the top 10 genetic PCs, whereas Model 2 also adjusted for education. Quantile–quantile (QQ) plots for the associations between the 116 AD risk SNPs and the 6 cognitive domain scores in Model 1 are shown in Supplementary Figure 4A. For most cognitive domain scores, the QQ plots indicate that the observed p values from the AD risk SNPs are more significant than expected by chance alone, indicating a potential excess of associations between the AD risk SNPs and measures of cognition.
Of the 116 AD risk SNPs, 1 SNP in CR1 (rs2093760) was associated with language/fluency at FDR q < 0.10 (Table 2) in Model 1. The association was attenuated but remained nominally significant (p < .05) after additional adjustment for education (Model 2). This SNP also was nominally associated with all other cognitive domain scores except orientation (Supplementary Table 7), and its MAF ranged from 0.066 to 0.107 across states/union territories (Supplementary Table 8). The other CR1 SNP investigated in this study, rs6656401, also had a nominal association with general cognitive function, language/fluency, memory, and visuospatial function (Supplementary Table 7).
Table 2.
SNPs With at least one Significant Association With Cognitive Domain Scores in LASI-DAD (FDR < 0.10), and APOE ε2 and ε4 Alleles
| SNP | Chr | Position | Nearest Gene | GWAS Study | Risk Allele | Risk AF in GWAS | Risk AF in LASI-DAD | Cognitive Measure | Model 1, Beta (p) | Model 2, Beta (p) |
|---|---|---|---|---|---|---|---|---|---|---|
| rs2093760 | 1 | 207786828 | CR1 | Jansen et al. (4). | A | 0.205 | 0.096 | Language/fluency | −0.18 (.001) | −0.14 (.003) |
| Effects of APOE alleles for comparison | ||||||||||
| rs429358 | 19 | 45411941 | APOE ε4 | N/A | C | 0.141 | 0.099 | General cognitive function | −0.06 (.366) | −0.03 (.544) |
| Orientation | −0.09 (.087) | −0.03 (.537) | ||||||||
| Executive function | −0.03 (.601) | −0.03 (.559) | ||||||||
| Language/fluency | −0.03 (.537) | −0.02 (.606) | ||||||||
| Memory | −0.06 (.388) | −0.04 (.458) | ||||||||
| Visuospatial function | −0.03 (.604) | −0.03 (.535) | ||||||||
| rs7412 | 19 | 45412079 | APOE ε2 | N/A | C | 0.928 | 0.954 | General cognitive function | −0.12 (.159) | −0.05 (.427) |
| Orientation | −0.07 (.361) | −0.02 (.726) | ||||||||
| Executive function | −0.09 (.310) | −0.05 (.437) | ||||||||
| Language/fluency | −0.18 (.013)* | −0.07 (.259) | ||||||||
| Memory | −0.12 (.248) | −0.09 (.281) | ||||||||
| Visuospatial function | −0.02 (.790) | 0.002 (.979) |
Notes: AF = allele frequency; Chr = chromosome; GWAS = genome-wide association study; LASI-DAD = Diagnostic Assessment of Dementia for the Longitudinal Aging Study in India; SNP = single-nucleotide polymorphism. Model 1 is adjusted for age, sex, and first 10 genetic PCs. Model 2 is adjusted for Model 1 covariates and education. Effect sizes (betas) are calculated with respect to the Alzheimer’s disease risk allele. Significant associations between single SNPs and cognitive domain scores at FDR < 0.10 are shown in bold font. Associations between APOE ε2 and ε4 alleles and cognitive domain scores at p < .05 are indicated by an asterisk.
For comparison purposes, Table 2 also shows the associations between APOE ε2 and ε4 with the cognitive domain scores. APOE ε4 was not associated with any of the cognitive domain scores, and APOE ε2 was associated only with language/fluency in Model 1 (p < .05). Aside from the association between APOE ε2 and visuospatial function in Model 2, beta coefficients for all APOE associations were in the expected direction, even if not significant. All associations for the 116 AD risk SNPs are shown in Supplementary Table 7. At a nominally significant p value (p < .05), additional 31 AD SNPs were associated with at least 1 cognitive measure (Model 1 or Model 2), with 19 SNPs associated in the expected direction. Among them, 10 SNPs (mapping to BIN1, CASS4, SORL1, ADAMTS1, PTK2B, and CR1) were associated with more than 1 of the 5 broad cognitive domain scores.
In contrast to the AD risk SNPs, p values from the general cognitive function SNPs generally follow a uniform distribution in both models for the cognitive domain scores (Supplementary Figure 4B). Only the QQ plots for visuospatial function exhibit slightly smaller p values than expected by chance alone. This suggests that most SNPs identified for general cognitive function from EA GWAS may not be associated with cognition in South Asians. Indeed, none of the 130 general cognitive function risk SNPs identified in EA were associated with cognitive domain scores in LASI-DAD after adjusting for multiple comparisons (Supplementary Table 9). However, 33 of the 130 SNPs were nominally associated with at least 1 of the 6 cognitive measures in Model 1 and/or Model 2. Among them, 21 SNPs were associated in the expected direction, where cognitive function–increasing alleles were associated with higher cognitive domain scores. Four of the 21 SNPs, which map to NMNAT2, HMGN4, MAPT, and RBL2, were associated with more than 1 of the 5 broad cognitive domains.
Association Between PRSs and Cognitive Domain Scores
We further assessed whether the candidate SNPs, when aggregated together as PRSs, were associated with cognitive measures. Briefly, 5 PRSs for AD were constructed from the AD GWAS in EA participants (3–7), after excluding variants in the APOE region. Note that there is an overlap in some of the SNPs that comprise these PRSs. The correlation between the 5 AD PRSs ranged from 0.50 to 0.80 (Supplementary Table 10). The PRS for general cognitive function (PRSGenCog) was created similarly, using the 130 candidate SNPs from the general cognitive function GWAS in EA (9). The correlations between PRSGenCog and the 5 AD PRSs ranged from 0.01 (with PRSAD_Bellenguez) to 0.06 (with PRSAD_Kunkle; Supplementary Table 10). We assessed whether each PRS was associated with each cognitive domain score separately using regression, with the same adjustment variables as Models 1 and 2 described earlier.
All associations between PRSs and cognitive domain scores were in the expected direction, with higher AD PRSs and lower general cognitive function PRS associated with lower cognitive function (Table 3). Specifically, all PRS(s) were associated with general cognitive function in Model 1 at p < .05. However, the associations of the AD PRS(s) and the cognitive function PRS seemed to be driven by different domains: AD PRS(s) were generally associated with executive function, language/fluency, memory, and visuospatial function with some variation by specific PRS, whereas the general cognitive function PRS was only associated with orientation. All associations attenuated in Model 2.
Table 3.
Association Between AD and General Cognitive Function PRSs and Cognitive Domain Scores in LASI-DAD
| PRSAD_Lambert | PRSAD_Jansen | PRSAD_Kunkle | PRSAD_Wightman | PRSAD_Bellenguez | PRSGenCog | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | p | Beta | p | Beta | p | Beta | p | Beta | p | Beta | p | ||
| General cognitive function | Model 1 | −0.057 | .033 | −0.061 | .021 | −0.081 | .002 | −0.052 | .048 | −0.054 | .039 | 0.059 | .027 |
| Model 2 | −0.042 | .046 | −0.041 | .055 | −0.058 | .006 | −0.027 | .203 | −0.041 | .051 | 0.031 | .143 | |
| Orientation | Model 1 | −0.022 | .331 | −0.017 | .451 | −0.037 | .104 | −0.020 | .388 | −0.016 | .487 | 0.072 | .002 |
| Model 2 | −0.013 | .545 | −0.005 | .818 | −0.021 | .301 | −0.005 | .827 | −0.008 | .693 | 0.056 | .007 | |
| Executive function | Model 1 | −0.051 | .055 | −0.057 | .031 | −0.071 | .006 | −0.050 | .059 | −0.056 | .033 | 0.044 | .095 |
| Model 2 | −0.037 | .081 | −0.037 | .085 | −0.050 | .020 | −0.025 | .243 | −0.043 | .044 | 0.017 | .439 | |
| Language/fluency | Model 1 | −0.047 | .033 | −0.059 | .007 | −0.056 | .010 | −0.036 | .102 | −0.024 | .273 | 0.020 | .351 |
| Model 2 | −0.037 | .059 | −0.047 | .017 | −0.041 | .036 | −0.021 | .281 | −0.017 | .402 | 0.005 | .799 | |
| Memory | Model 1 | −0.057 | .065 | −0.075 | .015 | −0.099 | .001 | −0.051 | .097 | −0.069 | .025 | 0.046 | .133 |
| Model 2 | −0.044 | .110 | −0.057 | .039 | −0.078 | .004 | −0.029 | .285 | −0.058 | .036 | 0.023 | .406 | |
| Visuospatial function | Model 1 | −0.071 | .009 | −0.046 | .093 | −0.091 | .001 | −0.063 | .020 | −0.062 | .022 | 0.040 | .148 |
| Model 2 | −0.059 | .015 | −0.030 | .216 | −0.072 | .003 | −0.044 | .070 | −0.052 | .031 | 0.019 | .440 |
Notes: AD = Alzheimer’s disease; LASI-DAD = Diagnostic Assessment of Dementia for the Longitudinal Aging Study in India; PRSAD_Lambert = Alzheimer’s disease polygenetic risk score calculated using the identified risk SNPs from Lambert et al. (3); PRSAD_Jansen = Alzheimer’s disease polygenetic risk score calculated using the identified risk SNPs from Jansen et al. (4); PRSAD_Kunkle = Alzheimer’s disease polygenetic risk score calculated using the identified risk SNPs from Kunkle et al. (5); PRSAD_Wightman = Alzheimer’s disease polygenetic risk score calculated using the identified risk SNPs from Wightman et al. (6); PRSAD_Bellenguez = Alzheimer’s disease PRS calculated using the identified risk SNPs from Bellenguez et al. (7); PRSGenCog = general cognitive function polygenetic risk score calculated using the identified risk SNPs from Davies et al. (9). Model 1 adjusted for age, sex, and first 10 genetic PCs. Model 2 adjusted for Model 1 covariates and education. All PRSs are standardized to an N(0,1) distribution. Associations significant at p < .05 are shown in bold font.
Variability of Cognitive Domain Scores Explained by Demographic and Genetic Factors
We next examined how much variability in the cognitive domain scores was explained by APOE ε2 and ε4, the 5 AD PRSs, and PRSGenCog individually or simultaneously. The percentage of variance in the cognitive domain scores explained by a base model of age, sex, and the first 10 genetic PCs ranged from 9.97% (visuospatial function) to 29.24% (language/fluency) (Table 4). APOE ε2 and ε4, which are known to be associated with AD, only explained an additional 0.03% (visuospatial function) to 0.49% (language/fluency) beyond the base model. The 5 AD PRSs explained an additional 0.04% (PRSAD_Bellenguez and orientation) to 1.11% (PRSAD_Kunkle and visuospatial function) beyond the base model, whereas PRSGenCog explained an additional 0.07% (language/fluency) to 0.77% (orientation). When the AD genetic risk factors were combined (5 AD PRSs and APOE ε2 and ε4), they explained 0.51% (executive function) to 1.39% (memory) over the base model. Further adding PRSGenCog to the model with the AD genetic risk factors resulted in a total of 1.11% (executive function) to 1.66% (memory) additional variance explained over the base model. When education was included, the genetic risk factors (5 AD PRSs, PRSGenCog, APOE ε2 and ε4) explained less variability, ranging between 0.45% (executive function) and 1.01% (memory).
Table 4.
Percentage of Variance in Cognitive Domain Scores Explained by Genetic Risk Factors in LASI-DAD
| General Cognitive Function | Orientation | Executive Function | Language/Fluency | Memory | Visuospatial Function | |
|---|---|---|---|---|---|---|
| Age + sex + 10 PCsa | 27.66% | 26.70% | 24.66% | 29.24% | 22.13% | 9.97% |
| APOEb | 0.21% | 0.28% | 0.10% | 0.49% | 0.16% | 0.03% |
| PRSAD_Lambertb | 0.36% | 0.08% | 0.30% | 0.35% | 0.29% | 0.66% |
| PRSAD_Jansenb | 0.42% | 0.05% | 0.38% | 0.56% | 0.50% | 0.28% |
| PRSAD_Kunkleb | 0.75% | 0.21% | 0.61% | 0.51% | 0.89% | 1.11% |
| PRSAD_Wightmanb | 0.31% | 0.06% | 0.29% | 0.21% | 0.23% | 0.53% |
| PRSAD_Bellenguezb | 0.33% | 0.04% | 0.37% | 0.09% | 0.43% | 0.51% |
| APOE + 5 AD PRSsb | 1.04% | 0.51% | 0.82% | 1.25% | 1.39% | 1.20% |
| PRSGenCogb | 0.39% | 0.77% | 0.23% | 0.07% | 0.19% | 0.20% |
| APOE + 5AD PRSs + PRSGenCogb | 1.53% | 1.36% | 1.11% | 1.37% | 1.66% | 1.46% |
| APOE + 5AD PRSs + PRSGenCogc | 0.65% | 0.75% | 0.45% | 0.83% | 1.01% | 0.86% |
Notes: AD = Alzheimer’s disease; APOE = apolipoprotein E; LASI-DAD = Diagnostic Assessment of Dementia for the Longitudinal Aging Study in India; PRS = polygenetic risk score; PRSAD_Lambert = Alzheimer’s disease polygenetic risk score calculated using the identified risk SNPs from Lambert et al. (3); PRSAD_Jansen = Alzheimer’s disease polygenetic risk score calculated using the identified risk SNPs from Jansen et al. (4); PRSAD_Kunkle = Alzheimer’s disease polygenetic risk score calculated using the identified risk SNPs from Kunkle et al. (5); PRSAD_Wightman = Alzheimer’s disease polygenetic risk score calculated using the identified risk SNPs from Wightman et al. (6); PRSAD_Bellenguez = Alzheimer’s disease PRS calculated using the identified risk SNPs from Bellenguez et al. (7); PRSGenCog = general cognitive function polygenetic risk score calculated using the identified risk SNPs from Davies et al. (9). All PRSs are standardized to an N(0,1) distribution.
aChange in R2 when the variable(s) were added to a null model.
bChange in R2 when the variable(s) were added to the following model: cognitive domain score ~ age + sex + 10 PCs.
cChange in R2 when the variable(s) were added to the following model: cognitive domain score ~ age + sex + 10 PCs + education.
Discussion
Genetic association studies in populations with complex ancestry are challenging, especially when GWAS from another ancestry, such as EA, are used to inform the selection of candidate SNPs/variants. Investigating the genetic associations in Indians/South Asians may be particularly challenging as they are racially, geographically, and genetically diverse. As discussed in the literature and demonstrated here, Indian/South Asian ancestry is characterized by a mixture of ASI and ANI, as well as many heterogeneous subpopulations due to stronger founder events (23,25,26,30,31). The extent of inbreeding, which is known to be associated with many traits including cognition (32), also varies substantially within and across the subpopulations. The complexity compounds because diverse genetic ancestry is also a product of cultural, historical, socioreligious, language, caste-based, and urban/rural influences in India (30). As a result, this population is likely to have a unique genetic architecture for cognitive function and AD, and operating genetic factors may also be influenced by a variety of social and cultural factors. Further, even though major efforts were put into training interviewers in different language groups, as well as analytically accommodating language differences in the construction of the cognitive factors, we found that significant proportions of variance in cognitive domain scores were explained by genetic ancestry. This may be due to causal genetic differences or may be a product of the changing sociocultural environment across the states and union territories that happens to track allele frequency clines.
Minor allele frequencies were different between LASI-DAD and the EA GWAS sample for about 80% of the SNPs we examined. Not surprisingly, SNPs identified in EA GWAS demonstrated limited transferability to LASI-DAD. However, our study highlighted 1 SNP in CR1 that may play an important role in cognitive function in Indians/South Asians. CR1, one of the first identified (3,33) and most widely studied AD risk genes, codes complement receptor 1 that is largely involved in glial-mediated inflammation and clearance of beta-amyloid (34). This gene is also associated with AD in East Asians (35,36), though different SNPs than those evaluated in this study show the strongest associations. Similar to what we observed previously (11), the APOE major isoforms (ε4 and ε2) have limited associations with cognitive function in this sample. APOE major isoforms are known to have a less pronounced effect on cognitive function than AD, and the strength of the association varies by instrument (37–41). In addition, the influence of APOE on cognition/AD might involve multiple variants and mechanisms in Indians/South Asians that differ somewhat from EA populations (e.g., interaction with other variants, local ancestry, or environmental factors) (11,42–45). All these reasons could contribute to the lack of association between APOE major isoforms and cognitive domain scores in this study. Future large-scale studies in Indians/South Asians with cognition and/or clinically diagnosed AD are needed to fully understand the function of APOE in this population.
Though we detected few associations after multiple testing corrections, most of the SNPs have effect directions that are consistent with those from the EA GWAS. Notably, there were also a handful of SNPs that were nominally associated in the unexpected direction. This could be due to a different LD structure in South Asian and EA populations such that the same risk allele tags different haplotypes (e.g., risk vs protective haplotypes) in each population. Similar heterogeneous associations have been observed in other studies. For example, the major allele (C) of SNP rs3865444, an AD risk allele in Europeans, was found to be protective in Han Chinese (46). Nonetheless, these associations were not significant after multiple testing corrections and should be interpreted with caution.
The lack of significant associations at the SNP level could be due to limited sample size and inadequate power, as we tested hundreds of SNPs with very small effect sizes. This notion is further supported by the observation that when those SNPs were aggregated together as PRSs, both the general cognitive function and AD PRSs were associated with general cognitive function in LASI-DAD, explaining up to 1.1% of the variance in cognitive measures in LASI-DAD. As a comparison, similar PRSs for general cognitive function explain between 2.6% and 4.3% of variance in general cognitive function in EA samples (9); however, the PRSs examined were constructed using genome-wide SNPs at various p value thresholds and thus are not directly comparable to the cognitive function PRS examined here. Several studies have examined the association between AD PRSs and cognition in EA, but these studies do not report the variance explained by the PRSs (8), which makes comparison across studies and ancestry groups difficult. The larger variance explained by AD PRSs than the general cognitive function PRS in LASI-DAD might be in part because AD is under stronger genetic control compared to cognition based on heritability estimates (1). Further, AD GWAS have a well-defined phenotype (clinical AD) whereas the general cognitive function GWAS investigated a measure of cognition that was defined differently across participating studies, potentially reducing the generalizability of the identified SNPs. Nonetheless, the amount of variance in the LASI-DAD cognitive domain scores accounted for by genetic risk factors is much smaller than that of nongenetic risk factors, especially education. This advocates an urgency to increase education equity across the country (47,48).
This study has some limitations. First, the sample size is small. Given the genetic heterogeneity of the population, a much larger sample size with greater participant diversity is critical to further investigate the genetic influences on cognition and dementia. Also, given that this genetic heterogeneity is likely confounded with educational, cultural, historical, socioreligious, language, caste-based, and urban/rural influences in India, stratified analysis by genetic and/or sociocultural subgroup would be an important next step in examining genetic associations. Second, we used the genetic data that were imputed to the 1000 Genomes Project reference panel, which lacks representativeness for Indian/South Asian populations (16,49). This may have introduced bias to the analysis, although we did not find evidence that it influenced the allele frequencies of the AD and general cognitive function risk SNPs per se. Large-scale whole-genome sequencing in Indian/South Asians would help to advance our understanding of the genetic factors that influence cognition in this population. Third, cognitive function in an older population may be influenced by many factors. Obtaining cognitive domain scores in specific population sub-groups (e.g., in participants with and without clinically diagnosed AD) could help strengthen the findings. Fourth, the cognitive tests were carried out in the mother tongues of the participants, including 12 local languages, which could potentially introduce bias. However, we performed vigorous validation and did not detect systematic bias associated with language (50). Given the rich diversity of geographical and social structures and the importance of social determinants of health for dementia (51–53), it will be critical to examine the interaction between genetics and age, sex, education, and/or other social determinants. Lastly, longitudinal studies with cognitive domain scores across multiple time points could further shed light on the potential roles of genetic factors in cognitive aging.
Although the extraordinary genetic complexity in Indian/South Asian presents analytical challenges, it also provides a unique opportunity to study disease-associated genetic variants that are more likely to be enriched in the presence of strong founder effects (26,54). The complex ancestral structure also allows us to study the effects of genetic variants in different local and/or global ancestral backgrounds. This can lead to the identification of new and unique variants not previously described. Moreover, studying genetic variation in this population can inform future efforts directed toward applying and testing new statistical methods that take into account complex ancestral structure.
In summary, our current study comprehensively describes the complex genetic structure of a sample of Indians/South Asians from multiple sampling locations across India. We also present the association between several cognitive domain scores and GWAS-identified SNPs for AD and general cognition function in this sample. Our study represents an important stepping stone toward better characterization of the genetic architecture of cognitive aging in the Indian/South Asian population and highlights the need for further research that may lead to the identification of new variants unique to this population.
Supplementary Material
Contributor Information
Wei Zhao, Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, Michigan, USA; Survey Research Center, Institute for Social Research, University of Michigan, Ann Arbor, Michigan, USA.
Jennifer A Smith, Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, Michigan, USA; Survey Research Center, Institute for Social Research, University of Michigan, Ann Arbor, Michigan, USA.
Yi Zhe Wang, Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, Michigan, USA.
Manjusha Chintalapati, Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, California, USA; Center for Computational Biology, University of California, Berkeley, Berkeley, California, USA.
Farah Ammous, Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, Michigan, USA.
Miao Yu, Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, Michigan, USA.
Priya Moorjani, Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, California, USA; Center for Computational Biology, University of California, Berkeley, Berkeley, California, USA.
Andrea Ganna, Institute for Molecular Medicine Finland, Helsinki, Finland.
Alden Gross, Johns Hopkins Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland, USA.
Sharmistha Dey, Department of Biophysics, All India Institute of Medical Sciences, New Delhi, India.
Joyita Benerjee, Department of Geriatric Medicine, All India Institute of Medical Sciences, New Delhi, India.
Prasun Chatterjee, Department of Geriatric Medicine, All India Institute of Medical Sciences, New Delhi, India.
Aparajit B Dey, Department of Geriatric Medicine, All India Institute of Medical Sciences, New Delhi, India.
Jinkook Lee, Department of Economics, University of Southern California, Los Angeles, California, USA.
Sharon L R Kardia, Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, Michigan, USA.
Funding
This project is funded by the National Institute on Aging (R01 AG051125, U01 AG065948). The study sponsor had no role in the design and conduct of the study; in the collection, analysis, and interpretation of the data; or in the preparation, review, or approval of the manuscript.
Conflict of Interest
None declared.
Author Contributions
Study concept and design: W.Z., J.A.S., P.M., J.L., S.L.R.K. Statistical analyses: W.Z., J.A.S., M.Y., Y.Z.W. Manuscript preparation: W.Z., J.A.S., M.Y., Y.Z.W., F.A., P.M., M.C. Data collection and study supervision: A.B.D., S.D., J.B., P.C., J.L., S.L.R.K. Critical revision of the manuscript: P.M., A.Ga., A.Gr., A.B.D., J.L., S.L.R.K.
References
- 1. Tucker-Drob EM, Briley DA, Harden KP. Genetic and environmental influences on cognition across development and context. Curr Dir Psychol Sci. 2013;22(5):349–355. doi: 10.1177/0963721413485087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scheltens P, Strooper BD, Kivipelto M, et al. . Alzheimer’s disease. Lancet. 2021;397(10284):1577–1590. doi: 10.1016/S0140-6736(20)32205-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. . Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45(12):1452–1458. doi: 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jansen IE, Savage JE, Watanabe K, et al. . Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51(3):404–413. doi: 10.1038/s41588-018-0311-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kunkle BW, Grenier-Boley B, Sims R, et al. . Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51(3):414–430. doi: 10.1038/s41588-019-0358-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wightman DP, Jansen IE, Savage JE, et al. . A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat Genet. 2021;53(9):1276–1282. doi: 10.1038/s41588-021-00921-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bellenguez C, Küçükali F, Jansen IE, et al. . New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat Genet. 2022;54(4):412–436. doi: 10.1038/s41588-022-01024-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harrison JR, Mistry S, Muskett N, Escott-Price V. From polygenic scores to precision medicine in Alzheimer’s disease: a systematic review. J Alzheimers Dis. 2020;74(4):1271–1283. doi: 10.3233/JAD-191233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davies G, Lam M, Harris SE, et al. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat Commun. 2018;9(1):2098. doi: 10.1038/s41467-018-04362-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alzheimer’s & Related Disorders Society of India. The Dementia India Report: prevalence, impact, costs and services for Dementia. (Eds) Shaji KS, Jotheeswaran AT, Girish N, Srikala Bharath, Amit Dias, Meera Pattabiraman and Mathew Varghese. New Delhi: ARDSI; 2010. [Google Scholar]
- 11. Smith JA, Zhao W, Yu M, et al. . Association between episodic memory and genetic risk factors for Alzheimer’s disease in South Asians from the Longitudinal Aging Study in India-Diagnostic Assessment of Dementia (LASI-DAD). J Am Geriatr Soc. 2020;68(Suppl 3):S45–S53. doi: 10.1111/jgs.16735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deary IJ, Corley J, Gow AJ, et al. . Age-associated cognitive decline. Br Med Bull. 2009;92:135–152. doi: 10.1093/bmb/ldp033 [DOI] [PubMed] [Google Scholar]
- 13. Harris SE, Deary IJ. The genetics of cognitive ability and cognitive ageing in healthy older people. Trends Cogn Sci. 2011;15(9):388–394. doi: 10.1016/j.tics.2011.07.004 [DOI] [PubMed] [Google Scholar]
- 14. Lee J, Banerjee J, Khobragade PY, Angrisani M, Dey AB. LASI-DAD study: a protocol for a prospective cohort study of late-life cognition and dementia in India. BMJ Open. 2019;9(7):e030300. doi: 10.1136/bmjopen-2019-030300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gross AL, Khobragade PY, Meijer E, Saxton JA. Measurement and structure of cognition in the Longitudinal Aging Study in India–Diagnostic Assessment of Dementia. J Am Geriatr Soc. 2020;68(Suppl 3):S11–S19. doi: 10.1111/jgs.16738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. 1000 Genomes Project Consortium, Auton A, Brooks LD, et al. . A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28(24):3326–3328. doi: 10.1093/bioinformatics/bts606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mallick S, Li H, Lipson M, et al. . The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538(7624):201–206. doi: 10.1038/nature18964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. GenomeAsia100K Consortium. The GenomeAsia 100K Project enables genetic discoveries across Asia. Nature. 2019;576(7785):106–111. doi: 10.1038/s41586-019-1793-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu C-C, Liu C-C, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9(2):106–118. doi: 10.1038/nrneurol.2012.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Corbo RM, Scacchi R. Apolipoprotein E (APOE) allele distribution in the world. Is APOE*4 a “thrifty” allele? Ann Hum Genet. 1999;63(Pt 4):301–310. doi: 10.1046/j.1469-1809.1999.6340301.x [DOI] [PubMed] [Google Scholar]
- 22. Patterson N, Moorjani P, Luo Y, et al. . Ancient admixture in human history. Genetics. 2012;192(3):1065–1093. doi: 10.1534/genetics.112.145037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moorjani P, Thangaraj K, Patterson N, et al. . Genetic evidence for recent population mixture in India. Am J Hum Genet. 2013;93(3):422–438. doi: 10.1016/j.ajhg.2013.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Purcell S, Neale B, Todd-Brown K, et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461(7263):489–494. doi: 10.1038/nature08365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakatsuka N, Moorjani P, Rai N, et al. . The promise of discovering population-specific disease-associated genes in South Asia. Nat Genet. 2017;49(9):1403–1407. doi: 10.1038/ng.3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tournebize R, Chu G, Moorjani P. Reconstructing the history of founder events using genome-wide patterns of allele sharing across individuals. PLoS Genet. 2022;18(6):e1010243. doi: 10.1371/journal.pgen.1010243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pemberton TJ, Absher D, Feldman MW, Myers RM, Rosenberg NA, Li JZ. Genomic patterns of homozygosity in worldwide human populations. Am J Hum Genet. 2012;91(2):275–292. doi: 10.1016/j.ajhg.2012.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kirin M, McQuillan R, Franklin CS, Campbell H, McKeigue PM, Wilson JF. Genomic runs of homozygosity record population history and consanguinity. PLoS One. 2010;5(11):e13996. doi: 10.1371/journal.pone.0013996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Majumder PP. The human genetic history of South Asia. Curr Biol. 2010;20(4):R184–R187. doi: 10.1016/j.cub.2009.11.053 [DOI] [PubMed] [Google Scholar]
- 31. Consortium GU, Sivasubbu S, Scaria V. Genomics of rare genetic diseases—experiences from India. Hum Genomics. 2019;14(1):52. doi: 10.1186/s40246-019-0215-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clark DW, Okada Y, Moore KHS, et al. . Associations of autozygosity with a broad range of human phenotypes. Nat Commun. 2019;10(1):4957. doi: 10.1038/s41467-019-12283-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lambert JC, Heath S, Even G, et al. . Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1094–1099. doi: 10.1038/ng.439 [DOI] [PubMed] [Google Scholar]
- 34. Johansson JU, Brubaker WD, Javitz H, et al. . Peripheral complement interactions with amyloid beta peptide in Alzheimer’s disease: polymorphisms, structure, and function of complement receptor 1. Alzheimers Dement. 2018;14(11):1438–1449. doi: 10.1016/j.jalz.2018.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma XY, Yu JT, Tan MS, Sun FR, Miao D, Tan L. Missense variants in CR1 are associated with increased risk of Alzheimer’ disease in Han Chinese. Neurobiol Aging. 2014;35(2):443.e17–443.e21. doi: 10.1016/j.neurobiolaging.2013.08.009 [DOI] [PubMed] [Google Scholar]
- 36. Zhang W, Jiao B, Xiao T, et al. . Association of rare variants in neurodegenerative genes with familial Alzheimer’s disease. Ann Clin Transl Neurol. 7(10):1985–1995. doi: 10.1002/acn3.51197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Deary IJ, Whiteman MC, Pattie A, et al. . Cognitive change and the APOE epsilon 4 allele. Nature. 2002;418(6901):932932. doi: 10.1038/418932a [DOI] [PubMed] [Google Scholar]
- 38. Small BJ, Rosnick CB, Fratiglioni L, Bäckman L. Apolipoprotein E and cognitive performance: a meta-analysis. Psychol Aging. 2004;19(4):592–600. doi: 10.1037/0882-7974.19.4.592 [DOI] [PubMed] [Google Scholar]
- 39. Jorm AF, Mather KA, Butterworth P, Anstey KJ, Christensen H, Easteal S. APOE genotype and cognitive functioning in a large age-stratified population sample. Neuropsychology. 2007;21(1):1–8. doi: 10.1037/0894-4105.21.1.1 [DOI] [PubMed] [Google Scholar]
- 40. Liu F, Pardo LM, Schuur M, et al. . The apolipoprotein E gene and its age-specific effects on cognitive function. Neurobiol Aging. 2010;31(10):1831–1833. doi: 10.1016/j.neurobiolaging.2008.09.015 [DOI] [PubMed] [Google Scholar]
- 41. Wisdom NM, Callahan JL, Hawkins KA. The effects of apolipoprotein E on non-impaired cognitive functioning: a meta-analysis. Neurobiol Aging. 2011;32(1):63–74. doi: 10.1016/j.neurobiolaging.2009.02.003 [DOI] [PubMed] [Google Scholar]
- 42. Agarwal R, Tripathi CB. Association of apolipoprotein E genetic variation in Alzheimer’s disease in Indian population: a meta-analysis. Am J Alzheimers Dis Other Demen. 2014;29(7):575–582. doi: 10.1177/1533317514531443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rajabli F, Feliciano BE, Celis K, et al. . Ancestral origin of ApoE ε 4 Alzheimer disease risk in Puerto Rican and African American populations. PLoS Genet. 2018;14(12):e1007791. doi: 10.1371/journal.pgen.1007791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Blue EE, Horimoto A, Mukherjee S, Wijsman EM, Thornton TA. Local ancestry at APOE modifies Alzheimer’s disease risk in Caribbean Hispanics. Alzheimers Dement. 2019;15(12):1524–1532. doi: 10.1016/j.jalz.2019.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Choi KY, Lee JJ, Gunasekaran TI, et al. . APOE promoter polymorphism-219T/G is an effect modifier of the influence of APOEε 4 on Alzheimer’s disease risk in a multiracial sample. J Clin Med. 2019;8(8):1236. doi: 10.3390/jcm8081236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tan L, Yu J-T, Zhang W, et al. . Association of GWAS-linked loci with late-onset Alzheimer’s disease in a northern Han Chinese population. Alzheimers Dement. 2013;9(5):546–553. doi: 10.1016/j.jalz.2012.08.007 [DOI] [PubMed] [Google Scholar]
- 47. Agrawal T. Educational inequality in rural and urban India. Int J Educ Dev. 2014;34:11–19. doi: 10.1016/j.ijedudev.2013.05.002 [DOI] [Google Scholar]
- 48. Asadullah MN, Yalonetzky G. Inequality of educational opportunity in India: changes over time and across states. World Dev. 2012;40(6):1151–1163. doi: 10.1016/j.worlddev.2011.11.008 [DOI] [Google Scholar]
- 49. Sengupta D, Choudhury A, Basu A, Ramsay M. Population stratification and underrepresentation of Indian subcontinent genetic diversity in the 1000 Genomes Project Dataset. Genome Biol Evol. 2016;8(11):3460–3470. doi: 10.1093/gbe/evw244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Banerjee J, Jain U, Khobragade P, et al. . Methodological considerations in designing and implementing the harmonized Diagnostic Assessment of Dementia for Longitudinal Aging Study in India (LASI-DAD). Biodemography Soc Biol. 2020;65(3):189–213. doi: 10.1080/19485565.2020.1730156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Russ TC, Stamatakis E, Hamer M, Starr JM, Kivimaki M, Batty GD. Socioeconomic status as a risk factor for dementia death: individual participant meta-analysis of 86 508 men and women from the UK. Br J Psychiatry. 2013;203(1):10–17. doi: 10.1192/bjp.bp.112.119479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vega IE, Cabrera LY, Wygant CM, Velez-Ortiz D, Counts SE. Alzheimer’s disease in the Latino community: intersection of genetics and social determinants of health. J Alzheimers Dis. 2017;58(4):979–992. doi: 10.3233/JAD-161261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. George KM, Lutsey PL, Kucharska-Newton A, et al. . Life-course individual and neighborhood socioeconomic status and risk of dementia in the Atherosclerosis Risk in Communities Neurocognitive Study. Am J Epidemiol. 2020;189(10):1134–1142. doi: 10.1093/aje/kwaa072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ankala A, Tamhankar PM, Valencia CA, Rayam KK, Kumar MM, Hegde MR. Clinical applications and implications of common and founder mutations in Indian subpopulations. Hum Mutat. 2015;36(1):1–10. doi: 10.1002/humu.22704 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.