Figure 1. Gliotransmission is required for axon regeneration.
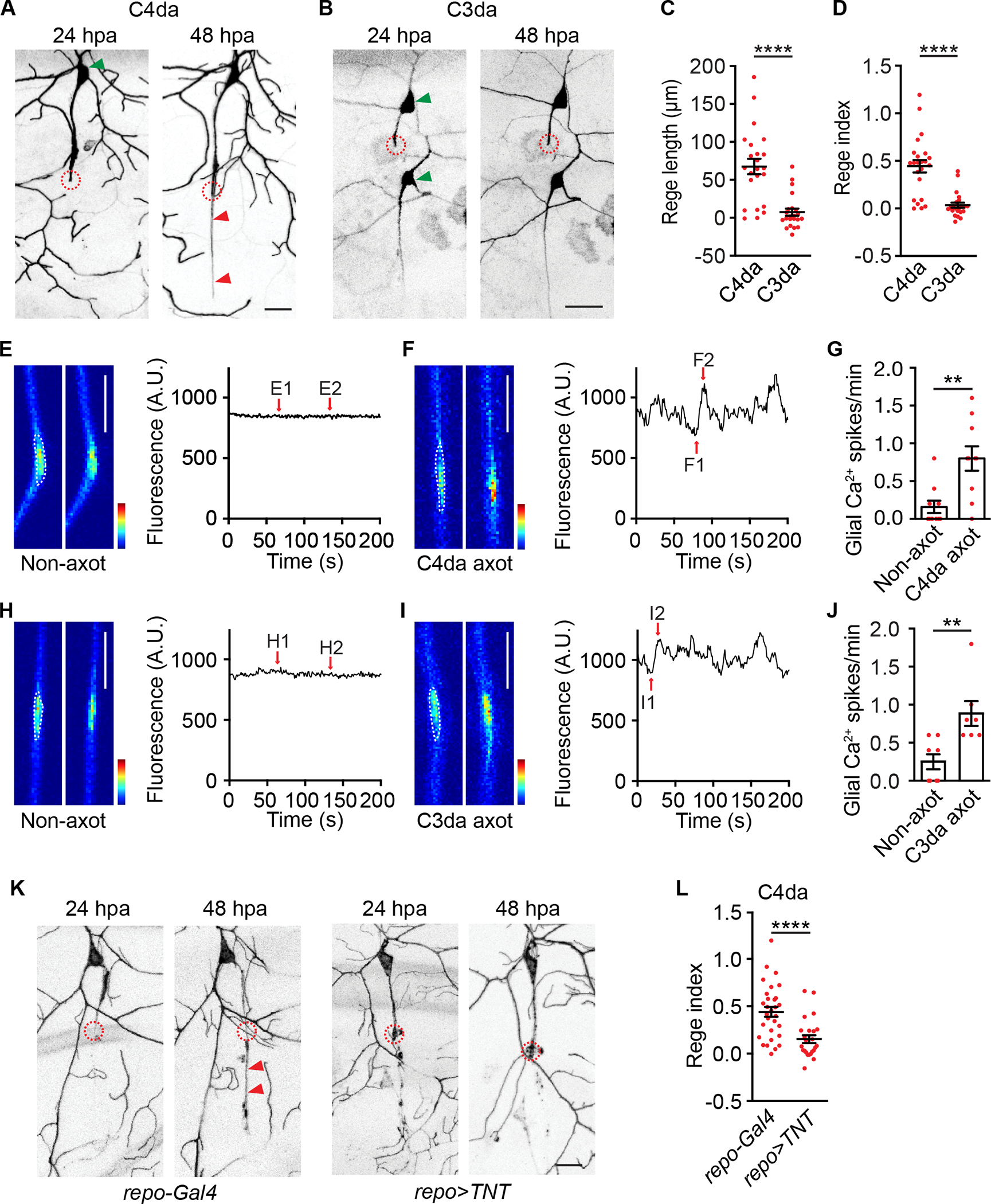
(A-B) Representative images of a C4da neuron (A) and a C3da neuron (B) at 24 and 48 hpa. Circles indicated axotomy sites. Red arrowheads in (A) indicated the regenerated axon, and green arrowheads indicated somata. ppk-CD4-tdGFP and repo-Gal80, 19-12-Gal4>UAS-EYFP labeled C4da and C3da neurons, respectively. Throughout this work, repo-Gal80 was used to suppress non-specific expression of 19-12-Gal4 in glia. Scale bar, 20 μm. Of the two C3da neurons in (B), axotomy was performed on the dorsal neuron ddaF (upper).
(C-D) Quantifications of axon regeneration length (C) and regeneration index (D). n=23 for C4da and n=22 for C3da neurons, respectively.
(E-G) Ensheathing glial Ca2+ spikes after axotomy of C4da neurons. Larvae bearing repo-Gal4>UAS-GCaMP6s, ppk-CD4-tdTomato were used. (E-F) Representative GCaMP6s images and traces showed glial Ca2+ signals without axotomy (E, non-axot) and at 24 hours after axotomy of C4da neurons (F, C4da axot). (G) Quantifications of glial Ca2+ spikes. Panels E1-E2 and F1-F2 showed individual frames. Glial Ca2+ spikes were quantified from regions indicated by white dashed lines. Scale bar, 20 μm. GCaMP6s color scale: 0–2,500. n=10 and 10.
(H-J) Analogous to (E-G), axotomy of C3da neurons induced Ca2+ spikes in ensheathing glia. Larvae bearing repo-Gal4>UAS-GCaMP6s, NompC-LexA>LexAop2-mCherry were used. Scale bar, 20 μm. GCaMP6s color scale: 0–3,000. n=8 and 7.
(K-L) Representative images (K) and axon regeneration index (L) of C4da neurons from control larvae (repo-Gal4 group: repo-Gal4, ppk-CD4-tdGFP) and larvae expressing TNT in glia (repo>TNT group: repo-Gal4>UAS-TNT, ppk-CD4-tdGFP). Circles indicated axotomy sites and arrowheads indicated the regenerated axon. Scale bar, 20 μm. n=30 and 24.
Mann-Whitney test (C, D, G, J and L). **P<0.01, ****P<0.0001.
