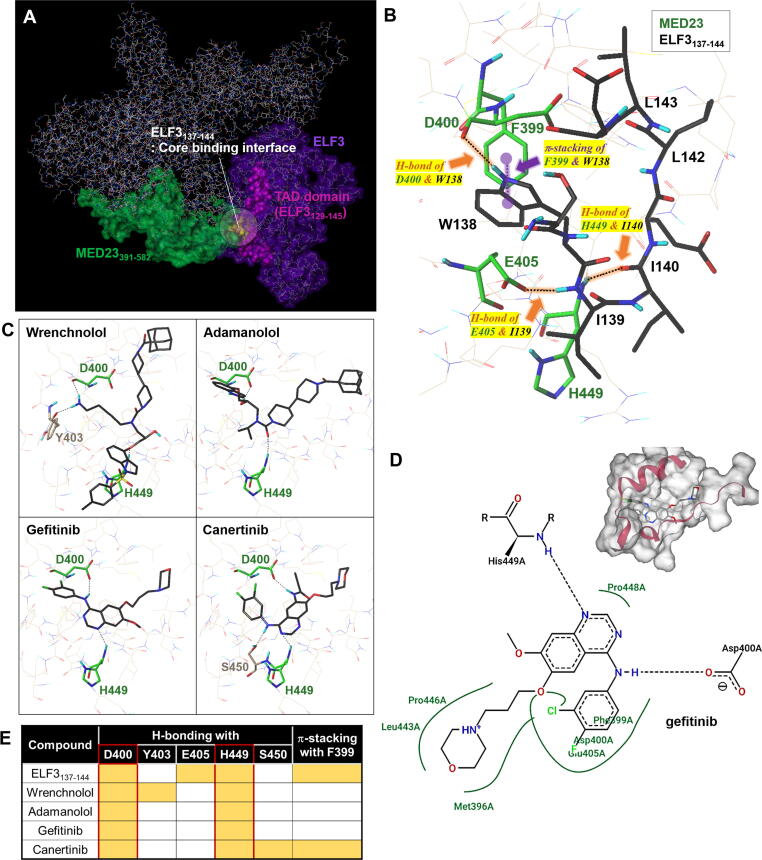
Fig. 1.
In silico-based structural analysis to predict hotspot of ELF3-MED23 complex. (A) Protein binding model of MED23WT and ELF3WT prepared using the Phyre2 server. The important residues responsible for ELF3–MED23 binding are depicted in different colors. (B) 3D docking view of the ELF3137-144 peptide on MED23 protein (PDB: 6H02). The key residues of MED23 and ELF3 involved in the interaction were labelled in green and black, respectively. The detailed 2D interaction diagrams between ELF3137-144 and MED23 is depicted in Figure S1A. (C) Docking pose of wrenchnolol, adamanolol, gefitinib, and canertinib that were previously identified to have ELF3-MED23 PPI inhibitory activity. (D) 2D interaction diagrams of the gefitinib docking generated by PoseView (https://proteins.plus). Gefitinib has been shown to have H-bondings to the D400 and H449 residues of MED23 with additional hydrophobic contacts via M396, F399, D400, E405, L443, P446, and P448 but no π-π interaction with F399. (E) Summary of H-bonding residues and -stacking of MED23 involved in the interaction with each of the indicated compounds. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)