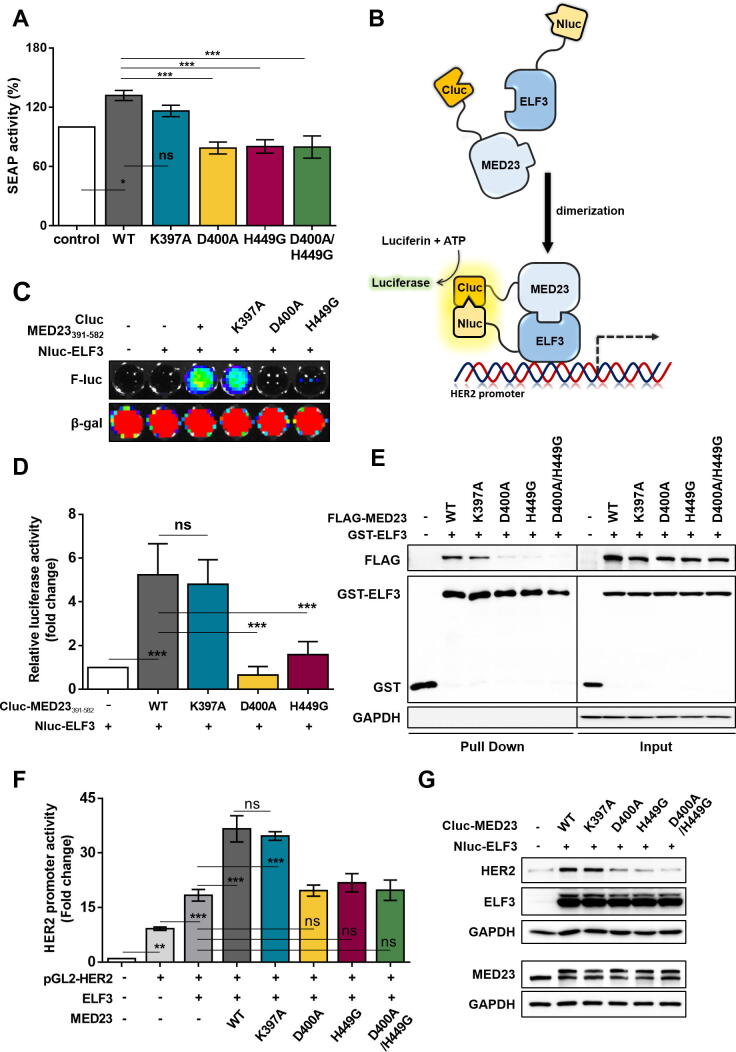
Fig. 2.
Specification of key residues and required interaction in the hotspot of ELF3-MED23 PPI. (A) To verify D400 and H449 of MED23 as key residues for ELF3-MED23 PPI, the impact of each MED23 mutant on the binding degree between gal4-ELF3 and MED23 was measured by SEAP activity. The results are expressed in bar graph (n = 5, mean ± S.D.). ANOVA (* and ***P < 0.05 and 0.001, respectively, ns = non-significant). MED23WT and MED23K397A increased the SEAP activity but D400A and H449G single mutants and D400A/H449G double mutant did not. (B) Graphical scheme of designed biosensor. The N-terminal fragment of luciferase was tagged to ELF3, and the C-terminal was linked to the MED23 constructs. (C, D) The degree of protein binding between ELF3 and different MED23391-582 fragments was evaluated by co-transfecting the generated constructs to HEK293 cells for 24 h. Representative image (C) and the quantification result (D) was demonstrated in a bar graph (n = 4, mean ± S.D.). ANOVA (***P < 0.001, ns = non-significant). (E) The extent of direct protein interaction between several full-length MED23 mutants and ELF3 was assessed by GST pull down assay. (F, G) Changes in the transcriptional activity (F) and HER2 expression level (G) were determined along with co-expression of ELF3 with different full-length MED23 constructs.