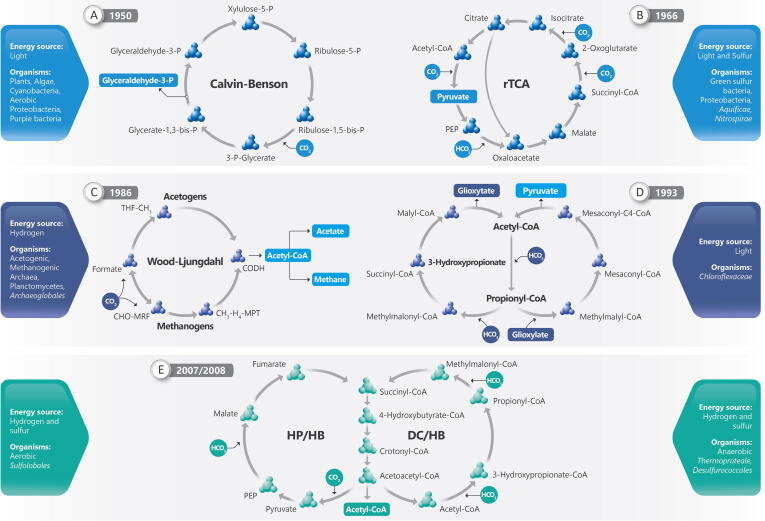
Fig. 2.
Natural carbon fixation. (A) CBB cycle; enzymes: ribulose-1,5-bisphosphate carboxylase/oxygenase, 3-phosphoglycerate kinase, glyceraldehyde-3-phosphate dehydrogenase, ribulose-phosphate epimerase. (B) rTCA cycle; enzymes: ATP-citrate lyase, malate dehydrogenase, succinyl-CoA synthetase, ferredoxin (Fd)-dependent-2-oxoglutarate synthase, isocitrate dehydrogenase, PEP carboxylase. (C) Wood–Ljungdahl cycle, upstairs acetogens Archaea and downstairs methanogens Archaea; enzymes: MPT-methylene tetrahydromethopterin, MFR-methanofuran, THF, tetrahydrofolate. (D) 3HP cycle; enzymes: acetyl-CoA carboxylase, propionyl-CoA carboxylase, methylmalonyl-CoA epimerase, succinyl-CoA:(S)-malate-CoA transferase, trifunctional (S)-malyl-CoA, -methylmalyl-CoA, mesaconyl-CoA transferase, mesaconyl-C4-CoA hydratase. (E) HP/HP and DC/HP cycle; enzymes: pyruvate synthase, PEP-carboxylase, malate dehydrogenase, fumarate hydratase/reductase, acetyl-CoA/propionyl-CoA carboxylase, 3-hydroxypropionate-CoA ligase/dehydratase, methylmalonyl-CoA mutase, succinyl-CoA reductase, 4-hydroxybutyrate-CoA ligase, crotonyl-CoA hydratase, acetoacetyl-CoA-ketothiolase.