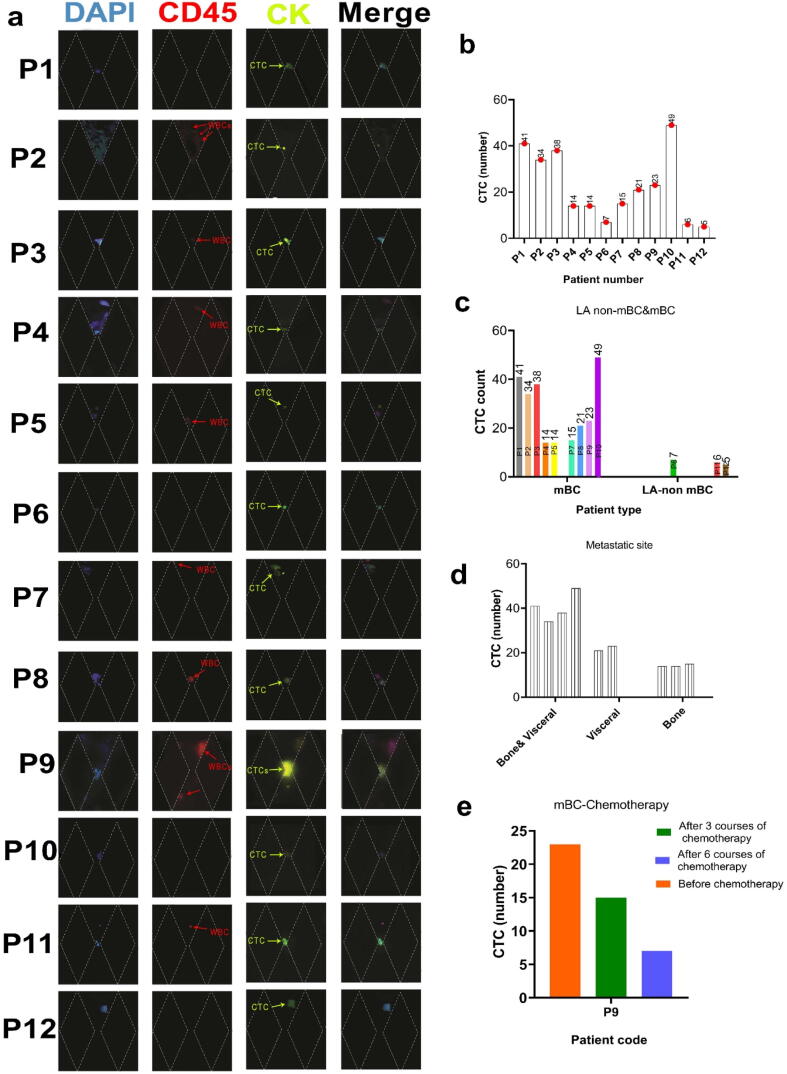
Graphical abstract.
Schematic of MPA-Chip.
Keywords: Circulating tumor cells, Microfluidics, Size separation, Cancer prognosis, Cancer therapy prediction
Highlights
-
•
Fabrication of micropillar array-based microfluidic chips with different geometries.
-
•
Conducting numerical simulations to compare velocity and pressure profiles.
-
•
Evaluating capture efficiency, purity and viability using various cancer cells.
-
•
Providing high capture efficiency (>85 %), purity and viability for CTCs isolation.
-
•
Clinical validation by counting CTCs in samples from patients with different breast cancer states.
Abstract
Introduction
The information derived from the number and characteristics of circulating tumor cells (CTCs), is crucial to ensure appropriate cancer treatment monitoring. Currently, diverse microfluidic platforms have been developed for isolating CTCs from blood, but it remains a challenge to develop a low-cost, practical, and efficient strategy.
Objectives
This study aimed to isolate CTCs from the blood of cancer patients via introducing a new and efficient micropillar array-based microfluidic chip (MPA-Chip), as well as providing prognostic information and monitoring the treatment efficacy in cancer patients.
Methods
We fabricated a microfluidic chip (MPA-Chip) containing arrays of micropillars with different geometries (lozenge, rectangle, circle, and triangle). We conducted numerical simulations to compare velocity and pressure profiles inside the micropillar arrays. Also, we experimentally evaluated the capture efficiency and purity of the geometries using breast and prostate cancer cell lines as well as a blood sample. Moreover, the device’s performance was validated on 12 patients with breast cancer (BC) in different states.
Results
The lozenge geometry was selected as the most effective and optimized micropillar design for CTCs isolation, providing high capture efficiency (>85 %), purity (>90 %), and viability (97 %). Furthermore, the lozenge MPA-chip was successfully validated by the detection of CTCs from 12 breast cancer (BC) patients, with non-metastatic (median number of 6 CTCs) and metastatic (median number of 25 CTCs) diseases, showing different prognoses. Also, increasing the chemotherapy period resulted in a decrease in the number of captured CTCs from 23 to 7 for the metastatic patient. The MPA-Chip size was only 0.25 cm2 and the throughput of a single chip was 0.5 ml/h, which can be increased by multiple MPA-Chips in parallel.
Conclusion
The lozenge MPA-Chip presented a novel micropillar geometry for on-chip CTC isolation, detection, and staining, and in the future, the possibilities can be extended to the culture of the CTCs.
Introduction
The isolation of circulating tumor cells (CTCs), which are found in the bloodstream of cancer patients [1], [2], [3], [4], is used to investigate the disease status of the patients, especially regarding metastasis [5], [6], [7]. It has been demonstrated that the presence or persistence of CTCs in cancer is correlated with recurrence, poor outcome, and resistance to therapy [8], [9]. The capture of CTCs from the peripheral blood may enable early assessments as well as genetic and pharmacological evaluation of cancer cells [10], [11]. Thus, isolation of CTCs is increasingly considered an essential part of the standard protocols for breast cancer diagnosis and treatment [12], [13]. However, the rarity of CTCs among blood cells (one in 109 blood cells) presents a technical challenge for capturing them, and reliable techniques are required to overcome this challenge [14].
Several techniques have been commercialized for capturing circulating tumor cells, such as the FDA-approved CellSearch® platform. This platform uses EpCAM antibody-coated magnetic particles, which capture a large number of white blood cells (WBCs) in addition to CTCs, thereby showing low efficiencies and purities [15], [16]. This is largely due to nonspecific WBC binding to the CTC-targeted antibody-coated magnetic particles [17]. Also, a recent study found that 19 % of primary tumors do not express EpCAM, and its expression is weak in another 35 %, suggesting that EpCAM-based technologies ignore a considerable set of EpCAM-negative CTCs in metastatic breast cancer [18]. Low precision of CTC and WBC sorting with poor specificity has limited the use of immune-capture micro-devices in CTC separation [19].
As an alternative, label-free methods use distinct physical properties of CTCs to separate them from WBCs instead of chemical affinity-based approaches [20], [21]. These methods do not depend on cell surface antigens and make it relatively straightforward to retrieve isolated cells since tumor cells do not bind to the surface of the micro device [20]. A variety of tag-free isolation methods are based on the physical properties of tumor cells such as size and deformability, membrane polarity, and acoustic behavior [22]. Among these methods, mechano-physical approaches use both the size and deformability of the cells for the capture mechanism. However, the relative coincidence of these properties between normal and cancerous cells lowers the yield, purity, and throughput of the isolation [23]. Using micropillar arrays (MPA) and deterministic lateral displacement (DLD) methods with controlled geometries, can alleviate this problem and improve the size-based entrapment of CTCs substantially [24], [25], [26], [27].
In the present study, to reach the maximum efficiency of CTC isolation and improve the capture purity, we studied the effect of the geometry and pattern of micropillars on cancer cell isolation. The main separation mechanism of MPA-Chip is the difference in deformation and size between leukocytes and red blood cells on the one hand, and CTCs on the other hand. As shown in Table S2 in the supplementary information, the size of normal circulating cells is generally smaller than 15 µm (WBCs: 7–15 µm; RBCs: 7–8 µm), and CTCs are generally>15 µm. We chose four micropillar geometries, i.e. triangle, circle, rectangle, and lozenge. We ran numerical simulations to compare velocity and pressure profiles inside the micropillar arrays. Also, we experimentally evaluated the capturing efficiency of the four geometries using breast and prostate cancer cell lines as representatives of CTCs. Moreover, capturing efficiency and purity were studied in cancer cell-spiked healthy blood samples for lozenge and rectangle shape micropillars. In addition, the device's performance was validated on 12 breast cancer (BC) patients at different stages. The lozenge chip presents a new optimized geometry as well as a new array of posts for on-chip CTC isolation, detection, and staining, and in the future, the possibilities can be extended towards the culture of the captured CTCs.
Materials and methods
MPA-chip design
Single-Chip design
The Single-Chip is composed of nine rows of posts; there are 21 posts in each odd row and 20 posts in the even rows. The distance between the posts for all shapes is reduced from 50 to 10 µm from the first row to the 9th row. Therefore, the distance between the obstacles reaches 10 µm in the last row. The height of the posts is 50 µm for all shapes. Table 1 displays the size and distances between each row for each shape.
Table 1.
Size and distances between rows for all micropillar shapes.
| Row number | Size (µm) |
|||||
|---|---|---|---|---|---|---|
| Distance gap between micropillars (µm) | Triangle (hypotenous- leg) | Circle (diameter) | Rectangle (length–width) | Lozenge (small-long diag.) | Distance micropillar center to micropillar center (horizontally) | |
| 1 | 50 | 50–39 | 50 | 50–30 | 50–200 | 100 |
| 2 | 45 | 55–41 | 55 | 55–30 | 55–200 | 100 |
| 3 | 40 | 60–43 | 60 | 60–30 | 60–200 | 100 |
| 4 | 35 | 65–44 | 60 | 65–30 | 65–200 | 100 |
| 5 | 30 | 70–46 | 70 | 70–30 | 70–200 | 100 |
| 6 | 25 | 75–48 | 75 | 75–30 | 75–200 | 100 |
| 7 | 20 | 80–50 | 80 | 80–30 | 80–200 | 100 |
| 8 | 15 | 85–52 | 85 | 85–30 | 85–200 | 100 |
| 9 | 10 | 90–54 | 90 | 90–30 | 90–200 | 100 |
Simulation
Numerical method
All shapes were designed in SOLIDWORKS® software and then implemented in COMSOL Multiphysics® simulation software. The governing equations of the blood fluid flow are the Navier-Stokes equation for the incompressible laminar flow, continuity, viscosity, and particle tracking.
| (1) |
| (2) |
Where P, μ and ρ are the pressure, dynamic viscosity,y and density of the fluid respectively. u is the velocity vector, t is time and g is the gravitational acceleration, and I is the identity matrix.
Blood is a non-Newtonian fluid, but it is represented in our simulations as a Newtonian fluid. The corresponding dynamic viscosity μ in Eq. (1) is calculated based on the non-Newtonian power law equation from the following equation.
| (3) |
And
| (4) |
with
| (5) |
Where m, n and are the fluid consistency coefficient, flow behaviour index, and lower shear rate limit respectively.
To verify the numerical method, each model was implemented 3 times under equal simulated conditions and the differences between the results obtained were examined. In the next step, we increased the input speed to the chip at a constant rate in 4 steps and evaluated the results. If the trend of the results is proportional to the rate of acceleration rate, the accuracy of the simulations performed is supported.
Simulation model
Fluid flow in microfluidic chips commonly is laminar because of their small scale and corresponding low Reynolds number. A non-compressible laminar flow simulation model is applied for blood fluid. Blood is a viscous and non-Newtonian fluid that can be described by the power viscosity equation Eq. (3). To implement the mathematical calculations, the computational fluid dynamics (CFD) method of COMSOL Multiphysics® 5.4.0.2 software was used.
For this simulation, 3-dimensional coordinates are applied to design the patterns. Initially, the desired patterns, which include circular, rectangular, triangular and lozenge barriers, were designed using SOLIDWORKS. The height of obstacles for all patterns was 50 µm. Then, the discussed patterns were entered into COMSOL. The governing equations for the patterns are non-Newtonian, non-compressible laminar flow (Laminar, Incompressible Flow Non-Newtonian) as well as particle tracking for fluid flow (Particle Tracing for Fluid Flow). The wall material is selected from dry and impermeable wall types. A free triangular mesh with a step of 0.1 µm is used; the number of mesh elements and total volume for each pattern is given in table S1 in the supplementary file.
Blood is a viscous fluid that begins to clot on contact with oxygen and its viscosity increases. Blood is a combination of 55 % plasma and 45 % cells. Cells include red blood cells, white blood cells, and platelets. The relative composition and physical properties of blood and cancer cell are shown in Table S2 in the supplementary file.
The solving method of direct PARADISO was applied for the phase initialization of laminar flow and time-dependent studies. The period of the simulation time was chosen from 0 to 5 s and time step was selected to be 0.1 ms. The velocity of blood was adjusted to a constant quantity while the pressure level at the outlet was set to zero.
Experimental
Microfluidic chip Fabrication
Soft lithography was used to fabricate microfluidic devices. The mold was created by patterning microfluidic channels on a silicon wafer by photolithography with SU-8 photoresist (MicroChem). Poly(dimethylsiloxane) (PDMS) was mixed with a cross-linker (Sylgard 184, Dow Corning) at a 10:1 ratio and then the mixture was poured on the mold, first it was degassed and then cured at 65 °C into the oven for at least 4 h. After the PDMS had cured, it was peeled off the mold, and the final device was built by bonding the PDMS to a glass substrate after surface activation with oxygen plasma. To prepare the fabricated devices, ethanol was flushed through the microfluidic channels and then washed with deionized water and PBS.
Cell culture and sample preparation
MDA-MB-231 and PC3 cells were harvested in a complete cell culture medium comprised of DMEM (Gibco Cat. no. 11995–065) with 10 % (vol/vol) FBS (Gibco Cat. no. 10099–133) and 1 % (vol/vol) penicillin–streptomycin (Gibco Cat. no. 15140–148) in T-25 cm2 flasks. Passages were performed when the cell confluence reached 70–80 %. Briefly, cells were washed with phosphate buffer saline (PBS) and were incubated with 1 ml Trypsin-EDTA solution (Gibco Cat. no. 25300–054) for 5 min. The cell suspension was centrifuged at 1200 rpm for 5 min and then the cell pellet was re-suspended in 2 ml fresh medium. The culture flask was kept at 37 °C in a cell culture incubator. MDA-MB-231 and PC3 cells with working concentrations of 500 × 103, 100 × 103, 1 × 103 cells per ml were used for optimizing capture efficiency and cell viability. Before experiments, PBS was used to prime the chip to prevent air bubbles from forming. To evaluate capture efficiency, MDA-MB-231 and PC3 cells were suspended in a 1 ml culture medium at concentrations of 105 and 100. Cell suspensions were injected into the devices using a syringe pump (0.5 ml/h), after 1 ml PBS was injected into the devices to wash the tubes and syringe. Fluid coming from the outlets was collected and cells were counted using lam neobar. The difference between the cell concentration at inlets and outlets was calculated and defined as capture efficiency. A trypan blue assay was utilized to evaluate the viability of captured cells. We injected 0.5 µL trypan blue to stain the captured dead cells inside the device and calculated the viability as the number of blue color cells per captured cells.
To evaluate the enrichment factor, 105 cells/ml from PC3 and MDA-MB-231 cells were suspended in the WBCs solution. The separation process was performed as mentioned in the capture efficiency (CE) section. The enrichment factor is defined as the ratio between the outlet concentration of cancer cells in WBCs and the initial concentration.
SEM sample preparation and imaging
Morphology and shape of device and micropillars were characterized using MIRA3 FESEM of TESCAN Company. In the cell culture section, the morphology of captured cells was studied by SEM Zeiss Supra 55VP. The cells were first fixed in 4 % paraformaldehyde and then cells were dehydrated in 60 %, 70 %, 80 %, and 90 % ethanol solutions in water and 100 % ethanol successively for 15 min for each step. The PDMS portion containing the captured cancer cells was separated from the glass substrate using a razor blade. Drying was conducted with a critical-point dryer (Tousimis Autosamdri-815), followed by sputtering (Cressington 208 HR) of the Pt/Pd coating on the samples.
Immunofluorescence staining
Cells were fixed using 4 % formaldehyde for 30 min for all chips and they were washed in PBS buffer (supplemented with 0.5 % BSA). In the following, cells were stained using a nuclear dye, 4′,6-diamidino-2- phenylindole (DAPI; Beyotime Institute of Biotechnology, USA) at 37 °C for 30 min. Also, white blood cells (WBCs) were stained by CD45 (1:100) antibody.
Blood sample preparation
The blood was collected via vein puncture from healthy human donors and stored in Vacutainers (BD) coated with EDTA as an anticoagulant. A sterile pipet was used to place 2 ml of Ficoll-Hypaque per milliliter of blood into a conical centrifuge tube with a 50-ml capacity. Anticoagulated blood was mixed with an equal volume of PBS. The diluted blood was slowly layered over the Ficoll-Hypaque solution by gently pipetting the diluted blood down the side of the tube containing the Ficoll-Hypaque. The tube was centrifuged for 30 min at 400 × g, 22 °C, with no brake. Mononuclear cells, or a desired non-aggregated mononuclear cell subset, were removed by pipetting sterile Pasteur across the interface between the plasma (upper layer) and the Ficoll-Hypaque (bottom). The aspirated mononuclear cells were transferred to a 15-ml conical tube. 10 ml PBS or tissue culture medium was added and mixed thoroughly. Then, the tube was centrifuged for 10 min at 400 × g, 4 °C. The supernatant was aspirated and the aggregated cells were redispersed in a culture medium of 2 ml. WBCs concentration was counted by Neubauer Haemocytometry. Regarding extracting tumor cell CE from the blood sample, Target cells were GFP-tagged to be used in the flow cytometry analysis. GFP-tagged MDA-MB-231 cells (100 × 103) were added to the prepared blood (Ficoll process, 5 × 106) and injection was performed using a syringe pump (flow rate: 0.5 ml/h). Isolation processes were the same as mentioned above and captured cells were imaged under a fluorescence microscope. A flow cytometry analysis of the WBC depleted from the target cells provided the CE of the isolation protocol from blood samples. A healthy blood test was conducted using the ethical code: IR.ACECR.REC.1396.1, which was taken from the ethics committee of the Motamed Cancer Institute, and consent letters were obtained from all participants before the study began.
Ethics statement
All experiments involving human study were conducted according to the ethical policies and procedures approved by the ethics committee of the Breast cancer research center, Motamed cancer institute, Iran (Approval no. IR.ACECR.REC.1396.1). Informed consent was obtained from all patients being included in the study.
Patient clinical study
The study was conducted at the Motamed cancer institute, Tehran, Iran. According to the National Health and Medical Research Councils’ guidelines, ethics approval was obtained from the breast cancer research center's Human Research Ethics Committee: IR.ACECR.REC.1396.1. Informed consent was obtained from 12 participants (breast cancer) to collect 10 ml of blood in K2E EDTA vacutainers. After the isolation process, CTCs on the chip were fixed with 4 % paraformaldehyde for 30 min. Cells were permeabilized using 0.2 % (v/v) Triton X- 100 for 10 min at room temperature, followed by blocking with 10 % FBS (Invitrogen) for 1 h at room temperature. In order to be classified as CTCs, CTCs must meet in visual inspection the following criteria: (1) pan-cytokeratin positive, (2) CD45 negative, (3) DAPI positive, (4) morphologically larger than background leukocytes, (5) intact nuclei. The results were reported as the number of CTCs per 10 ml whole blood.
Statistical analysis
GraphPad Prism, version 9.1.2 (GraphPad Software, USA) was employed for the statistical analysis. We considered p < 0.05 to be statistically significant.
Results
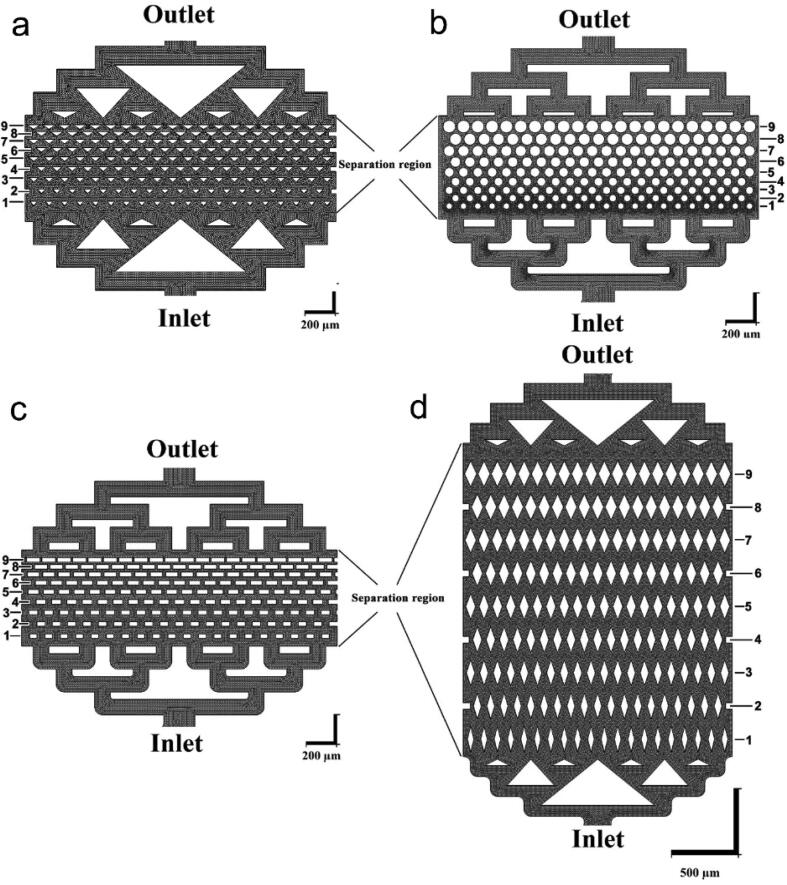
Fig. 1(a-d) shows schematic diagrams of the MPA chips with different pillar geometries. Each chip has an inlet, a separation region, and an outlet. The working fluid, cell suspension in culture medium or blood with CTCs, is injected into the chips from the inlets. After the inlet, the fluid flow splits into multiple streams and enters the micro-pillar area with 9 rows of micro-pillars. The gap between the micro-pillars decreases gradually from 50 µm in the first row to 10 µm in the last row. The height of the posts, and therefore the height of the micro-channels, is 50 µm in all the devices.
Fig. 1.
Schematic images of MPA-chip with micro-pillars with the triangle (a), circle (b), rectangle (c) and lozenge (d) cross-sections. The fluid flow enters the chip at the inlet and exits from the outlet. The black region indicates the fluid area and the white region shows the pillars and other solid parts of the chip. Every chip contains 9 rows of micro-pillars (horizontal in these images) in the separation region; the numbers next to each image indicate the row numbers of the micro-pillars. The gap between the pillars decreases from 50 µm in the first row to 10 µm in the last row.
Design of the MPA-Chip
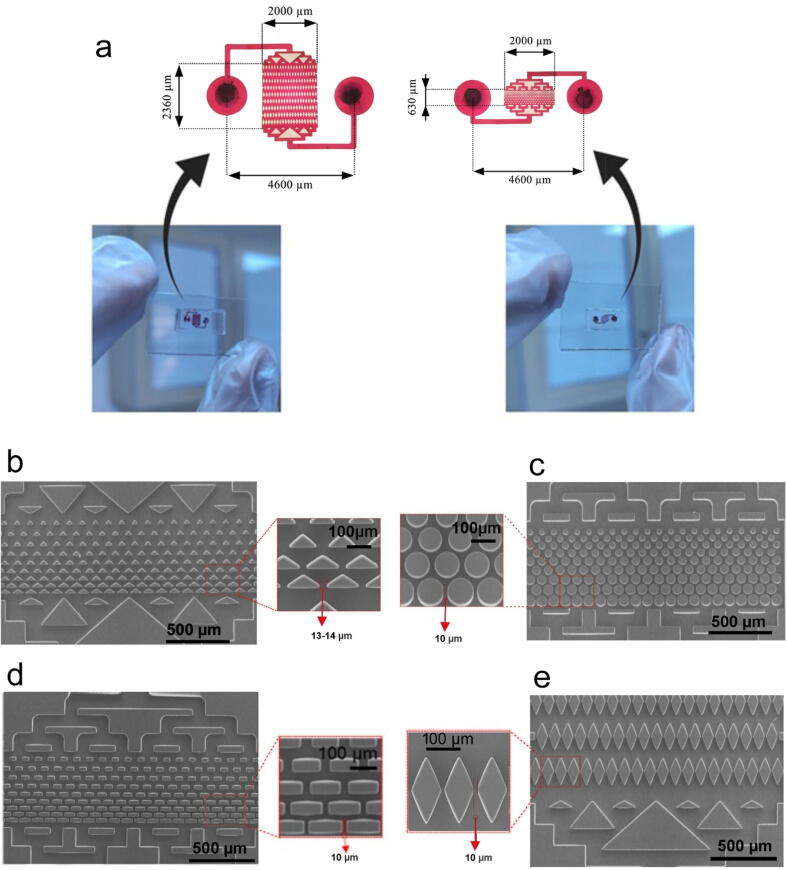
Four different geometries, i.e. triangle, circle, rectangle and lozenge, were chosen for the fabrication of the devices. Existing CTC separation devices often use large amounts of raw materials to fabricate the chips and stain the cells and have a large imaging area to visualize trapped cells [28]. We minimized the dimensions of the chips and volume of the microchannels to decrease the costs of the chip microfabrication process and reagents, especially the antibodies used in the experiments. As shown in Fig. 2a, the total size of the chips was 5.5 mm × 4 mm (lozenge) and 5.5 mm × 3 mm (triangle, rectangle, circle). Also, the size of the separation region and the volume of the MPA-chips were 0.63 mm × 2 mm for the triangle, rectangle, circle and 2.36 mm × 2 mm for the lozenge, respectively, and the total chip volume was smaller than 0.45 µL. Due to the small size of the chip, we were able to fabricate a SU-8 mold with approximately 130 chips on a single 4-inch silicon wafer.
Fig. 2.
(a) Pictures of MPA-chips including their dimensions. (b-e) SEM images of devices with different micropillar geometries; (b) triangle, (c) circle, (d) rectangle and (e) lozenge.
As shown in Fig. 2(b–e), scanning electron microscopy (SEM) of the fabricated micropillars confirmed that all the pillars had their designed geometry and size (with a 10 µm gap between micropillars) except for the triangular shape. The corners of the triangular pillars were too sharp (60°) to be reproduced exactly during fabrication. This resulted in slightly round corners and consequently larger distance between the pillars (approximately 13 µm).
Micropillar pattern and geometry determine the flow velocity and pressure in the MPA-Chip
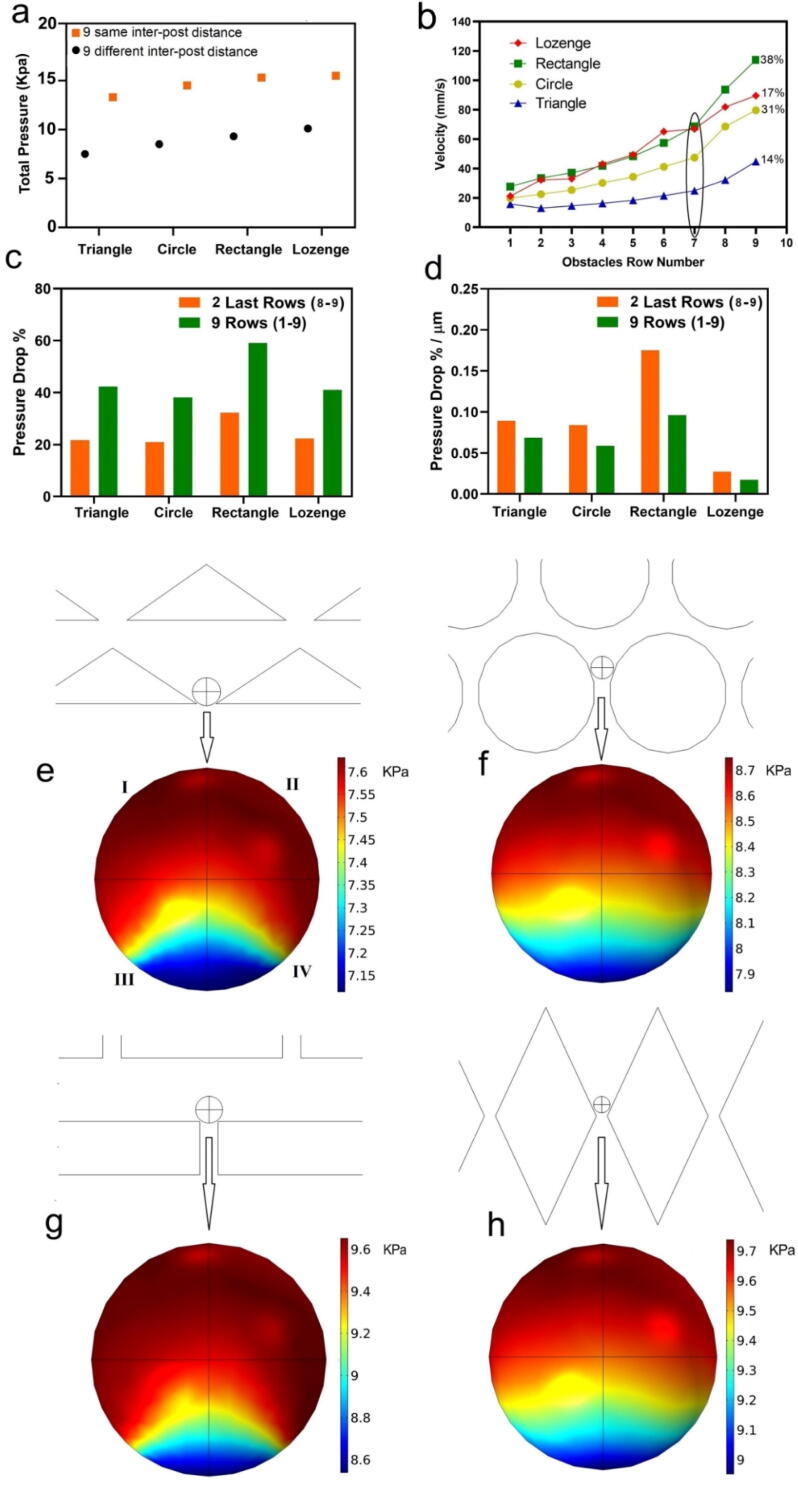
The geometry of micropillars and their arrangement in CTC devices determine the flow velocity and pressure in the chips [29]. From previous studies, we know that the velocity and pressure of the flow between the pillars have a large influence on the capture efficiency, purity and viability of the trapped cancer cells [30]. To understand the effect of pillar geometry on these parameters, we chose triangles, representing sharp surfaces not aligned with the flow direction, circles, representing curved surfaces, rectangles, representing flat surfaces, and lozenges which represent sharp surfaces aligned with the flow direction (Fig. 1). To investigate whether the arrangement of pillars in the chip has an effect on the cells we used COMSOL Multiphysics simulations. As shown in Fig. 3 (a), we found that the total pressure drop over the chip was higher when the inter-pillar distance was the same for all 9 rows than for the design in which the inter-pillar distance gradually decreased with pillar row number. Accordingly, in this state, the pressure acting on the trapped cells is larger, and there is a higher risk of cell rupture and lower cell viability. Therefore, we designed the MPA-chip with gradually decreasing gaps between the micropillars, from 50 µm in the first row to 10 µm in the last row, as shown in Fig. 1. By gradually reducing the gaps between micropillars, the total pressure drop over all chips is decreased by approximately 40 % (Fig. 3a), which likely results in better pressure distribution and a higher chance of trapped cells survival.
Fig. 3.
Results of numerical simulations of fluid pressure and velocity in the MPA-chip with different geometries. (a) The total pressure drop over the chip when the distance between the pillars is the same for all the 9 rows (10 µm) or different (from 50 µm in the first row to 10 µm in the 9th row.) (b) The fluid velocity magnitude through the rows of the MPA-Chip, measured between the micropillar; the plot also includes the calculated the percentage of fluid velocity increase in the last two rows (after the 7th row) of the chip compared to the velocity increase over the whole chip (subtraction of outlet velocity from inlet velocity) for all geometries. The flow rate of the inlet blood was 2.5 ml/h. (c) Percentage of pressure drop over the last 2 rows and over all the 9 rows for all the geometries compared to the pressure drop over the whole chip (subtraction of outlet pressure from inlet pressure) for all geometries. (d) Pressure drop percentage per micrometer over all 9 rows and over the last 2 rows for all pillar geometries compared to the pressure drop over the whole chip (subtraction of outlet pressure from inlet pressure) for all geometries. (e-h) fluid pressure acting on trapped particles (15 µm) in the chip for (e) triangle (f) circle (g) rectangle (h) lozenge.
Furthermore, we analyzed the flow velocity and pressure in the MPA-Chip to compare these parameters between different micropillar array geometries. We assumed blood as the fluid in the chip, a flow rate of 2.5 ml/h and zero pressure at the outlet. Fig. 3 (b-d) show the velocity and pressure in different rows of the chips with different geometries. The velocity increased from the first to the last row of the pillar geometries. This increase was especially sharp after row 7, and 14 %, 31 %, 38 % and 17 % of the total velocity increase happened after row 7 for the triangle, circle, rectangle and lozenge micropillars, respectively. The total increase in fluid velocity was the smallest for triangular pillars and the largest for the pillars with rectangular shape as seen in Fig. 3(b). The fluid velocity increased for each subsequent row of micropillars as the gap between them decreased, so that the flow-through area decreased while the flow rate was constant. Since the gap size in each row was similar between different micro-pillar geometries, the difference between velocity magnitudes of the different MPA-Chips reflects the net effect of the geometry type. Additional simulation results of the velocity for each geometry are shown in Figure S1 of the supporting information.
Fig. 3(c) shows the percentage of pressure drop over all the 9 micro-pillar rows as well as over the last 2 rows (rows number 8 and 9) compared to the pressure drop over the whole chip. As shown in this figure, the pressure drop was dependent greatly on the geometry of the pillars, and like the velocity, the pressure changed significantly after the last 2 rows (decreasing by around 20 % of the total pressure drop for the triangle, circle and lozenge and 32 % for rectangle micro-pillars) (Fig. 3(c)). Since the length of the separation region was larger for the chip with lozenge micro-pillars, we calculated the pressure drop per unit length in the MPA-Chip (Fig. 3(d)). Specifically, the chip with lozenge micropillars showed a significantly lower value compared to the other geometries (Fig. 3(d)), which is understandable since the total streamwise length of the lozenge chip is the largest (see Fig. 1). Also, a small difference between the pressure drop per unit length over all 9 rows and over the last 2 rows in the chip indicates the uniformity of the pressure drop within the chip. As a result, according to Fig. 3(d), the pressure drop is nearly uniform in the whole chip, in all geometries except for the rectangle micropillars in which there is a large pressure drop over the last 2 rows compared to other rows. The uniformity of the pressure drop over the separation area ensures that blood cells and CTCs are evenly distributed between the layers and that the pressure applied on them is uniform (See Movie S1 and S2). Additional simulation results of pressure (heat map and pressure distribution data) for each geometry are shown in Figure S2. (a and b) in the supplementary file. Also, the velocity and pressure distribution curves (Particle sizes: 15, 20 and 25 µm) are shown in Figures S1 and S2 in the supporting information, and Movies S1 and S2 in the supporting information show the particle distribution based on the flow field. Also, in order to compare the simulations with the experiments and in particular, the effect of red and white blood cells on the distribution of cancer cells, we carried out two different simulations. In the first, 1 µl of blood fluid is considered as the base fluid including RBCs, WBCs, and platelets according to the concentrations given in Table S2, and 100 cancer cells were also included. In the second simulation, only 100 cancer cell are added in the blood plasma, without any other cells. The results (videos) for both simulations are presented in Movie S1 and Movie S2 in the supporting information, respectively. The movies show that blood fluid and cancer cells are distributed uniformly throughout our chip.
During CTC separation, when cells are trapped between the micro-pillars, the fluid flow applies pressure on the surface of the cells and compresses them against the walls. If the pressure is too high, it can decrease the cell viability as well as the capture efficiency and if it is too low, it does not trap the cells with the intended purity. To compare the pressure levels on the trapped particles between the chips with different geometries, we simulated the flow over the particles. We considered 100 particles with a diameter of 15 µm representing CTCs and calculated the maximum pressure difference over the trapped particles as well as the pressure distribution (specifically its uniformity) on their surface according to the color contour plots in Fig. 3 (e-h). The maximum pressure difference on the particles is defined as the difference between the pressure at the top and bottom points (Fig. 3 (e-h)) on the particle. The maximum pressure difference was 0.45, 0.8, 1, and 0.7 kPa for the devices with triangle, circle, rectangle, and lozenge micro-pillar geometries, respectively. The results show that rectangle chips induce a larger pressure difference over the trapped particles. Furthermore, in order to calculate and compare the pressure distributions on the trapped particle in all the geometries, we defined a high-pressure area as that in which the value of the pressure falls within the top 15 % of the total pressure range in the trapped particle; this top 15 % pressure area is colored red in Fig. 3(e–h). The results show that about 75 % of the surface area of trapped particles between rectangular pillars experienced high pressure (red areas) whereas this was about 55 % for the triangle and 40 % for both circle and lozenge designs. To quantify the effect of pressure on the particles, we multiply, for each micropillar geometry, the maximum pressure difference with the high-pressure area percentage, so that a representative force is obtained. This results in (0.45 × 55 %) = 24.75 %, (0.8 × 40 %) = 32 %, (1 × 75 %) = 75 %, and (0.7 × 40 %) = 28 % for the devices with triangle, circle, rectangle, and lozenge micro-pillar geometries, respectively. In conclusion, the flow pushes the trapped particles between rectangular micro-pillars with a larger pressure over a larger surface area, i.e. a larger effective force, compared to the other geometries.
A comparison of different microarray geometries performance: Capture efficiency, enrichment, and cell viability
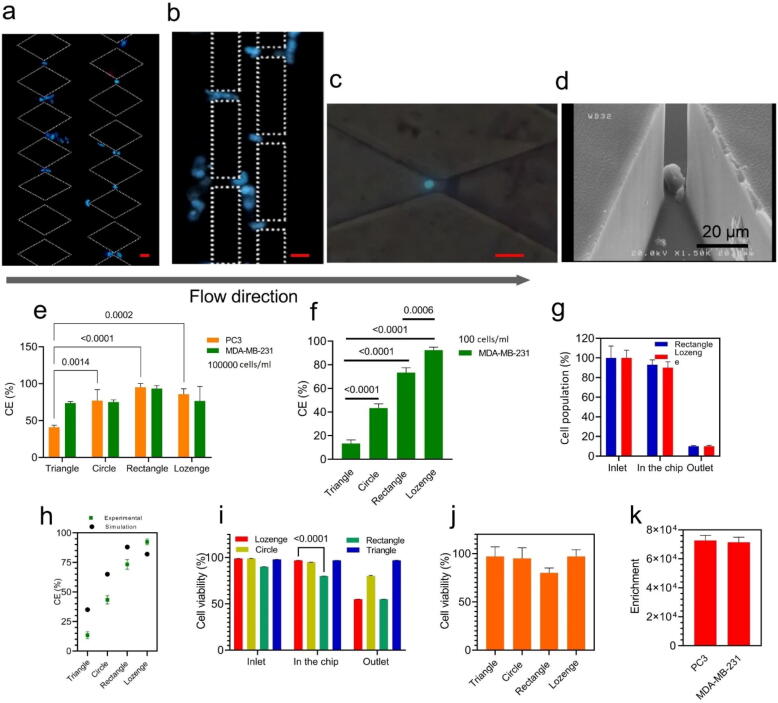
CTC isolation has a number of performance characteristics, including capture efficiency, enrichment factor, and cell viability [31], [32]. To verify that the fabricated chips can selectively separate the CTCs based on their size for different cancer types, we tested the performance of our micro-devices using two cancer cell lines: PC3, a human prostate cancer cell line and a highly invasive [33], [34], and MDA-MB-231, a human breast cancer cell line, and highly invasive [35], [36]. The DAPI-stained MDA-MB-231 cells (105cells/mL) trapped within the rectangle and lozenge devices are shown in Fig. 4 (a-c). The cancer cells were mainly trapped at the end of the separation area where the distance between the micropillars was smaller than the CTC size (<15 µm). The DAPI stained cells for other geometries are shown in Figure S4.
Fig. 4.
Characterization of device performance with different cancer types, where the cells are suspended in a culture medium. (a-c) Nuclei staining of captured cells (DAPI) inside the rectangle and lozenge chips in the last two rows (8th and 9th rows); scale bars are 20 µm in all the images. (d) SEM image of captured cells inside a device with lozenge micro-pillars. (e) Capture efficiency for two cell lines, human prostate cancer cell line PC3 and breast cancer cell line MDA- MB-231 (cell concentration: 105 cells/mL in culture medium). (f) Capture efficiency for MDA-MB-231 cells (100 cells/mL in culture medium). (g) The observed cell population (100 MDA-MB-231 cells suspended in 1 ml culture medium) flowing in various regions of the devices with lozenge and rectangular micro-pillars. Almost all cells entered the chip (observed at “inlet”) and only few cells were counted at the “outlet”; hence, cells were mostly captured in the separation region of chips and the rate of cell loss in the tubes and syringes at the inlet and outlet was very low and not significant. (h) Comparison of the simulation and experiment capture efficiency data for all geometries with 100 microparticles (representing cancer cells in the simulations, size: 15 µm) and 100 cells (MDA-MB-231, in the experiments), respectively. (i) Cell viability percentage for MDA-MB-231 cells in the inlet, on the chip and outlet and (j) viability of captured cells for PC3 cells only in the chip (i&j: cell concentration: 105 cells/mL in culture medium). (k) The enrichment factor of the chip for PC3 and MDA-MB-231 cells spiked into the same concentration of WBCs (5 × 105 cells/mL in culture medium). For all figures, all columns and plots represent the mean value and the standard deviation (n = 4).
We tracked the DAPI stained MDA-MB-231 cells inside the chip, and as shown in the movie in the supplementary file (Movie S3), DAPI stained cancer cells entered from the inlet and easily passed through the separation region’s primary arrays and were finally trapped in the last rows. Fig. 4 (d) shows an SEM image of a single cell that is captured by lozenge micropillars. We also observed cell clusters that were trapped inside the chip using SEM in Figure S5 (supplementary file).
In the following, we evaluate the capture efficiency (CE) of different geometries using high and low CTC concentrations (105 and 102 cells) in a 5 ml culture medium. The cancer cells, MDA-MB-231 and PC3, had a mean diameter of 15.5 µm, and 18.1 µm, respectively [37], [38]. As shown in Fig. 4 (e), for the high cell concentration, the rectangle and lozenge micropillars captured 92 ± 5 % and 85 ± 6 % of spiked CTCs respectively, while the circle and triangle micropillars isolated 60 ± 13 % and 43 ± 4 %, respectively. The CE was similar for MDA-MB-231 and PC3 cells (Fig. 4(e)).
Moreover, we investigated the CE for 100 MDA-MB-231 cells dispersed in 1 ml culture media. Lozenge and rectangle geometries separated 92 ± 3 % and 73 ± 3 % of cancer cells, respectively (Fig. 4(f)). These results for circle and triangle geometries were 41 ± 2 % and 16 ± 2 %, respectively. Therefore, we found no strong dependency of CE on cell concentration since we observed similarly high CE for lozenge and rectangle geometries when the concentration of spiked cells reduced from 105 to 102 cells. We tracked cells in all parts of the devices (inlet, separation region, and outlet) to monitor where our fabricated devices could physically capture cells. Fig. 4 (g) shows the number of cells we observed flowing in the respective areas (relative to the initial cell number), and clearly cells were mostly captured in the separation region of chips and the rate of cell loss in the tubes and syringes at the inlet and outlet was very low and not significant. Therefore, our results show that the separation region and whole chip worked well and efficiently. To verify the numerical method, the experimental and simulation data in the capture efficiency were evaluated. Firstly, using COMSOL Multiphysics, we simulated 100 microparticles with an average size of 15 µm representing 100 MDA-MB-231 cells for calculating capture efficiency (Fig. 4 (h) and Movie S2). Experimental and simulation results on capture efficiency (CE) show similar trends: according to Fig. 4 (h), the lozenge geometry has the highest CE in both the simulation and the experiment, namely 84 % and, 92 % respectively.
Then, for a better understanding of how micropillar geometry influences the viability of the isolated cells, we evaluated the cell viability in the three sections of devices (inlet, inside the chip, and outlet) for MDA-MB-231 cells. Fig. 4 (i) shows that the viability in the inlet is the same for all the geometries and close to 100 %, but not inside the chip or near the outlet. Hence, the cell viability does depend on the geometry. The viability of the cells inside the chip was over 95 %, except for the rectangular pillars (around 80 %). In the outlets, cells in the rectangular chip had a low cell viability of just around 48 %, while the cell viability in the other chips exceeded 75 %. The viability of PC3 cells is presented in Fig. 4 (j); this is similar to MDA-MB-231 for all the micro-pillar geometries.
The enrichment factor is an indication of how effective a filtration method enriches a specific target cell within a cell suspension in the presence of other cell types. To calculate this parameter, we used the ratio of tumor cells to leukocytes (white blood cells), and divided the ratio after filtration by the ratio before the filtration by the chip [39], [40]. We evaluated the enrichment factor of the chip for PC3 and MDA-MB-231 cells spiked into a culture medium along with the same concentration of WBCs. The measured enrichment factors were 7.3 × 104 for PC3 and 7.1 × 104 for MDA-MB-231 (Fig. 4 (k)). This result indicates that the enrichment factors are similar between PC3 and MDA-MB-231 cells, which agrees with results obtained by other studies in the field of size-based separation of CTCs [39], [41].
The best geometry for isolating CTCs?
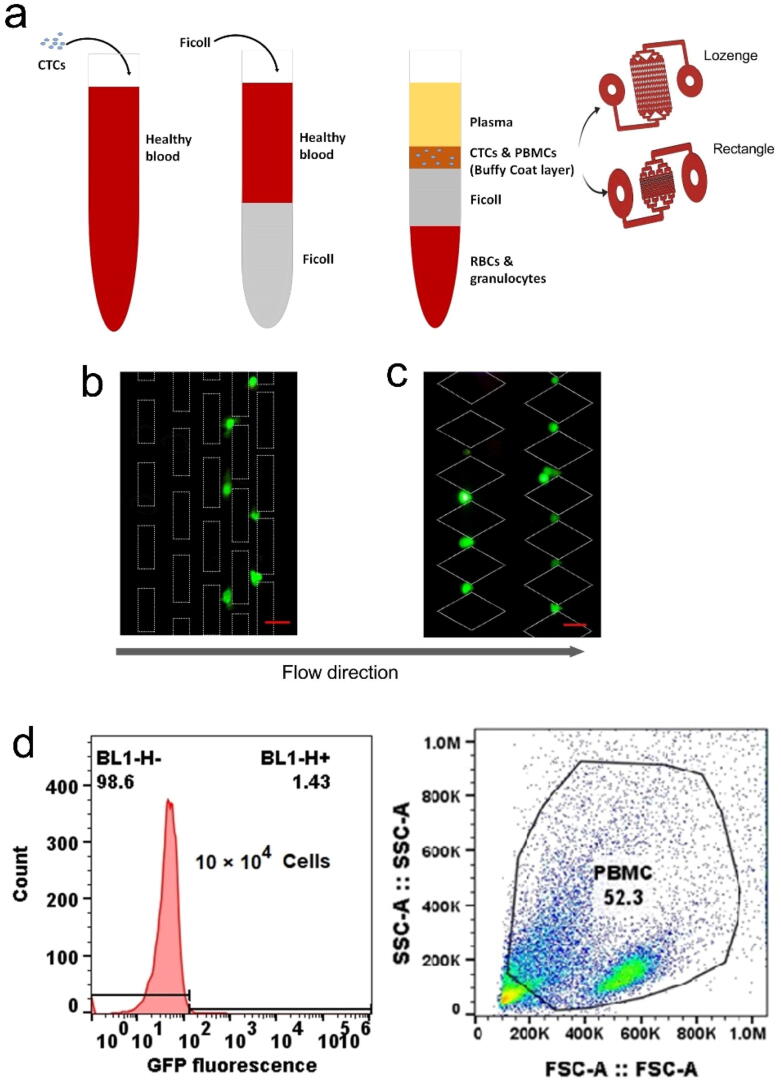
According to previous sections, both our simulations and experiments show that the rectangle and lozenge geometries are the most efficient geometric shapes for capturing CTCs. Therefore, we selected these two types for isolating CTCs from whole blood. Firstly, as shown in Fig. 5 (a), using the Ficoll technique, the buffy coat layer which contained the CTCs was separated and red blood cells (RBCs), platelets and granulocytes were depleted from the blood for improving the isolation and detection process and reducing clogging and process time [42]. Then, to characterize the device with blood samples, 105 GFP-tagged MDA-MB-231 human breast cancer cells were first spiked artificially into 5 ml unlabelled blood samples from healthy donors and applied to the MPA-Chip (Fig. 5(a)). In order to determine capture efficiencies, fluorescent microscopy and flow cytometry were used to image and count the GFP-tagged cancer cells that were captured. As shown in Fig. 5 (b,c), single cells of GFP-tagged MDA-MB-231 were trapped between the micropillars, which can also isolate cancer cell clusters (Figure S5). Then, we calculated the capture efficiency (CE) for the lozenge and rectangle geometries using flow cytometry (Figure (d-f)). The flow cytometer is a powerful tool technology in detecting and counting cells for validating the obtained CE of CTCs in blood samples [43]. Fig. 5(d,e) demonstrate flow cytometry histograms of 105 GFP-tagged cancer cells spiked in healthy donor blood for the lozenge (Fig. 5 (d)) and the rectangle design (Fig. 5(e)), respectively. The difference between the population of GFP + cells in the inlet and the outlet of the devices divided by the GPF + cells in the inlet represents the CE. The CE of lozenge and rectangular micropillars was 87 ± 3 % and 89 ± 2 %, respectively as represented in Fig. 5 (f). The calibration curve of different numbers of (105, 104, 103, and 102) GFP-tagged cancer cells spiked in healthy donor blood is demonstrated in figure S6. Moreover, we evaluated the purity of the separated cancer cells from a healthy blood sample by flow cytometry, by counting the population of WBCs before and after the isolation process (Fig. 5(g)). The percentages of non-captured WBCs for the rectangular and the lozenge designs were 95.1 ± 3.7 % and 91.5 ± 4.5 % respectively. This result demonstrated the population of WBCs captured with CTCs simultaneously and for lozenge and rectangular pillars was less than 10 %.
Fig. 5.
Characterization of the lozenge and rectangle chips with spiked breast cancer cell line (MDA-MB-231) in whole blood. (a) Schematic illustration of CTC isolation process in the blood sample, (b) and (c) Trapped GFP-labelled MDA-MB-231 human breast cancer cells (green) between micro-pillars in the last rows of the chip (8th and 9th rows); scale bar is 20 µm. (d) and (e) Flow cytometer histograms of GFP-labelled MDA-MB-231 cells obtained after collecting the liquid suspension coming out of the chip for the rectangle and lozenge chips, respectively. (f) Capture efficiency of the trapped GFP-labelled MDA-MB-231cells spiked into whole blood for lozenge and rectangle chips using flow cytometry. (g) Purity of the separated MDA-MB-231 cells in the population of prepared healthy blood on the chip. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
CTC counts in different states of breast cancer patients
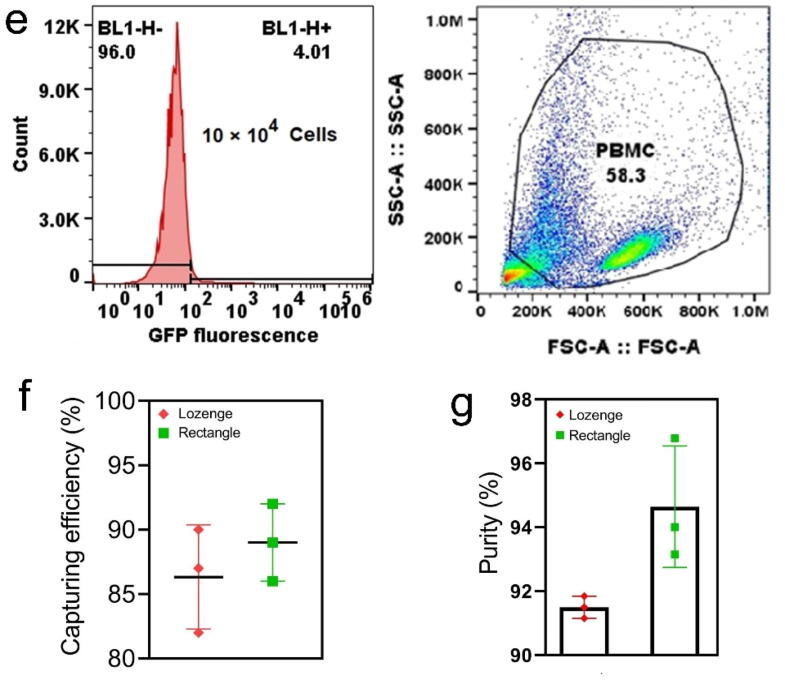
To clinically evaluate the MPA-chip with lozenge micropillar geometry, blood samples from twelve breast cancer patients were utilized to detect and count CTCs. First, 10 ml of each patient’s blood was incubated with the Ficoll to remove RBCs and some WBCs and then the buffy coat layer was suspended in a 1 ml MDEM cell culture medium. Then, samples were injected into our chip and analysed. The isolated cells on the chip were stained by DAPI, CD45, and pan-CK immunologically [9], [44]. CTCs were defined as DAPI+, CD45−, CK+ cells with high nucleus to cytoplasmic ratio and morphologically larger than background leukocytes, whereas WBCs were DAPI+, CD45+, and CK− [44].
In this study, we assessed the CTC counts in blood samples of 12 breast cancer patients in different states of disease. The patients were categorized into two groups: locally advanced non-metastatic breast cancer (LA non-mBC) and metastatic breast cancer (mBC). The clinical characteristics of patients are depicted in Table 2.
Table 2.
Clinical and pathological characteristics of breast cancer patients.
| Clinical characteristics | |
|---|---|
| Number of Patients (n) | 12 |
| Age(years) | |
| Mean | 60 |
| Range | 43–78 |
| Gender | |
| Male | 1 |
| Female | 11 |
| Metastatic sites | |
| Visceral | 2 |
| Bone | 3 |
| Bone & Visceral | 4 |
| Tumor stage | |
| Locally Advanced non-metastatic Breast Cancer (LA non-mBC) | 3 |
| Ⅳ-metastatic Breast Cancer (mBC) | 9 |
| HER-2 | |
| Positive | 3 (25 %) |
| Negative | 9 (75 %) |
Fig. 6(a) shows captured CTCs between micropillars in the MPA-chip. CTCs were captured from the blood samples of all the 12 patients, with a minimum and maximum concentration of 5 and 49 CTCs in 10 ml whole blood, respectively (Fig. 6(b)). The locally advanced non-metastatic breast cancer group (LA non-mBC) as shown in Fig. 6c had a mean number of 6 CTCs while the patients in the metastatic breast cancer (mBC) group displayed a mean number of 22 CTCs. Thus, CTC levels in metastatic disease were significantly higher than non-metastatic breast cancer patients (p\0.001). In the group of metastatic breast cancer (mBC; n = 9), 4 patients harbored both bone and visceral metastases, 3 bone metastases, and 2 visceral metastases involvement only. As shown in Fig. 6(d), patients with both visceral and bone lesions showed the highest median CTC count of about 49, whereas patients with only visceral lesions showed a median CTC count of 23. For patients with only bone disease, the median CTC number was 14 which was significantly lower compared to the group with both visceral and bone metastases (p = 0.0005) and to only visceral metastatic patients (p = 0.001) (Fig. 6(d)).
Fig. 6.
Detection and enumeration of circulating tumor cells (CTCs) in breast cancer patients. (a) Fluorescent images showing detected CTCs from patient samples. Cells were stained by DAPI (blue), CK (green arrow, CTCs), and CD45 (red arrow, WBCs); CTCs are DAPI+, CK+, and CD45−; scale bar represents 20 μm. (b) The numbers of CTCs detected from blood samples of 12 breast cancer patients; results are from 10 ml whole blood. (c) The number of CTCs detected in venous blood samples from non-metastatic breast cancer with locally advanced tumors (non-mBC; n = 3), and from metastatic breast cancer (mBC; n = 9). (d) Comparison of the number of CTCs according to their metastatic niche with those with only soft tissue involvement (n = 3), only bone metastases (n = 3), or both soft tissue and bone metastases (n = 3). (e) CTC numbers before and after different courses of chemotherapy. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
CTC counts for one of the mBC patients was also calculated after 3 and 6 courses of chemotherapy. As the values are shown in Fig. 6 (e), the number of CTCs that was 23 before treatment decreased after 3 and 6 courses of chemotherapy to 15 and 7, respectively. (Fig. 6 (e)).
Discussion
Although various geometries have been used in the literature for size and deformability-based CTC capture, there has been no comprehensive comparison between micropillar geometries. In this study, we demonstrated a comprehensive comparison of four basic micropillar geometries relevant to CTC isolation in both simulation and experiment. Our simulations focused on two main parameters: the velocity profile and the pressure profile in the chip. As mentioned before, the fluid velocity between the micro-pillars and the pressure drop over the last rows of the micropillar array plays a significant role in capturing the cells. Abrupt changes in the pressure drop, such as the changes shown in the rectangular pillar geometry (see Fig. 3 (c,d)), may result in a strong pressure acting on the cells trapped in the micropillar gaps and consequently in a decrease of the cell viability of the capture cells, which in turn compromises the capturing efficiency.
Based on the simulations and experiments, Table 3 provides an overview of the results to facilitate the discussion and analysis. Accordingly, the lozenge chip had lower ΔP (pressure drop over the last two pillar rows) and ΔV (velocity difference between the first and last pillar row) than the rectangle chip. Since the rectangular geometry effectively forms a wall-like obstacle against the flow of fluid in the last two rows, we observed the highest velocity between the gaps for this geometry [45], [46]. Correspondingly, we observed lower capture efficiency (for 100 cancer cells) for the rectangular geometry than for the lozenge geometry. The pressure drop over the last rows of the MPA-chip affects the viability of trapped cells. In the rectangular geometry, higher ΔP and high pressure applied to a large area of the captured cell surface (75 % of its surface) reduced the cell viability (also see Fig. 3 (g) and Fig. 4 (i)). In the chips with rectangular micro-pillars, the larger pressure and velocity can also create a force on the cells that pushes them through the trap. In the lozenge chips, the pressure drop and velocity are lowest, and so the cell viability and the CE are the highest. Pressure-driven deformability for metastatic cells such as MDA-MB-231 has been previously reported [47]. The effect of pressure and shear stress on the deformability, migration, and viability of CTCs has been widely investigated [48], [49]. Congnart et al. reported that shear stress and mechanical constrictions induce morphological and molecular changes in the MDA-MB-231 cells [50]. The effect of shear stress on the viability of the tumorigenic cell lines was also investigated by Hyler et al. [51]. According to these previous reports, pressure-driven mortality may be caused when the cells are trapped between micro-pillars [50], [51]. The higher velocity between the gaps and the corresponding higher shear stress in the rectangle chips, especially in the last arrays, may be further increased when the cells are entrapped between the micropillars as shown in Fig. 3 (g). Hence, our simulation results (pressure and velocity variations) and the experimental results (viability, CE) are consistent.
Table 3.
An overview of simulation (ΔP and ΔV) and experimental results (CE and cell viability) of the MPA-chips with different geometries.
| Geometry | ΔP(KPa)* | ΔV (mm/s)# | CE×(%) | Cell viability (%) |
|---|---|---|---|---|
| Rectangle | −0.0961 | 86.20 | 73.3 ± 2 | 80 |
| Lozenge | −0.0174 | 68.34 | 92.3 ± 3 | 97 |
| Circle | −0.0587 | 59.89 | 41 ± 2 | 95 |
| Triangle | −0.0687 | 28.98 | 16 ± 2 | 97 |
ΔV is defined as the difference in velocity between the first and last row.
ΔP is defined as the pressure drop over the two last rows.
CE is the calculated capture efficiency (CE) for 100 cells.
Lozenge and rectangle micropillars successfully isolated CTCs with the highest efficiency (CE > 90 %) compared with triangles and circles. Fatih Sarioglu and co-workers [24] developed a microfluidic device consisting of triangle shape pillars for CTC cluster isolation. They demonstrated that a microchip with triangle pillars with 12 µm gaps physically captured 99 % of only MDA-MB-231 clusters, and no single cells. Zhou et al. reported a microfluidic device with diamond shape pillars (gap size: 12 µm) that showed 70 % trapping efficiency for single or cluster cells [28]. In this study, we obtained a capture efficiency of about 93 % for lozenge chips. Also, some studies have reported that in physical and size-based cell separation microchips, the array of triangular micropillars had a higher capture efficiency compared to circular micropillars [52], [53]. In this study on the other hand, we observed that the circular geometry had a higher capture efficiency compared to triangle geometry. The reason for this could be due to our new chip design, which gradually reduces gaps between micropillars in consecutive rows, which improves pressure distribution according to simulation results (Fig. 3).
For simulating a liquid biopsy and real conditions, we spiked MDA-MB-231 cells into healthy blood samples. In the lozenge and rectangle chips, these samples showed high capture efficiency (>85 %) and purity (>90 %), in contrast to several reported size-based technologies that display efficiencies of 50–70 % [54], [55].
Commercially available Vortex chips (Vortex Biosciences) show high purity (57–94 %) but low efficiency (up to 37 %) when used with diluted blood and spiked MCF7 breast cancer cells [56]. ParsortixTM (ANGLE) [57] and CellseeTM (Celsee Diagnostics) [58] devices have been found to achieve capture efficiency rates up to 62.5 % when using prefixed or diluted blood samples loaded with prostate or breast cancer cells with minimal leukocyte contamination (>80 %). The Parsortix and whole blood provided capture efficiencies of 70 %, however, only for large size cells (>20 μm) [59]. It is noteworthy that all systems described above require sample post-processing and also have difficulty discriminating CTCs from WBCs, often resulting in poor isolation or a high level of WBC contamination. A comprehensive comparison of efficiency, purity and other parameters affecting the separation of CTCs in this study with other DLD devices is shown in Table S3 (supplementary file). The capturing efficiency and purity of our device are very high compared to previous studies, but ours has no antibodies and no post-processing. The separation mechanism of MPA-Chip is based not only on the difference in size between normal circulating cells (leukocytes and RBCs) and CTCs, like in DLD approaches, but also on their difference in deformability. So, it has the potential to be developed into a product with relatively low costs, which at the same time is user-friendly, enhancing its usability in the clinic. The MPA-chip does still have limitations in volume capacity per unit of time (0.5 ml /h) compared to some studies but, due to the use of the Ficoll method, the blood volume is decreased from 7.5 ml to 0.5 ml without wasting the CTCs. The complete processing time for the isolation and detection of CTCs was 2 h. Also, the chip throughput can be increased by using multiple MPA-Chips in parallel. Thus, this method improves the system and allows it to analyze larger volumes of whole blood in a shorter period of time by reducing disturbing factors and the volume capacity.
Our MPA-chip with lozenge micropillar geometry was also validated with the detection of CTCs in blood samples from 12 breast cancer patients. As demonstrated in this work, we obtain a clear relationship between CTCs count and breast cancer progression, since metastatic patients show significantly higher CTCs numbers compared to non-metastatic metastatic. Therefore, in our study, patients with higher CTCs represent more advanced states of disease with higher recurrence risks. Chen et al. [60] found that the median number of CTCs in a metastatic group (8 cells/5 ml) was significantly higher than in the non-metastatic group (2 cells/5 ml; P < 0.001). Moreover, Cristofanilli et al. [61] evaluated CTC enumeration for the staging of metastatic breast cancer patients. Their results show that patients with Stage IVindolent disease had lower CTC numbers and longer overall survival in comparison to patients with Stage IVaggressive disease [61]. In the context of prospective clinical trials, CTC count can be considered an important tool for the staging of advanced disease and disease stratification.
There is an association between metastatic sites and the heterogeneity of breast cancer. Our result show that patients with only bone metastases displayed the lowest CTCs level, whereas the highest CTC count was found in the group of both bone and visceral metastatic patients. The patient group with only visceral metastasis showed a slightly elevated CTC count compared to the group with only bone metastasis. In addition, we observed that the amount of CTCs decreased as the chemotherapy courses continued suggesting that the treatment protocol was effective (Fig. 6 e). Accordingly, this device can be utilized for monitoring treatment results and defining the best treatment choice for patients. Yu et al. [62] showed that the incidence of CTCs in patients’ blood with visceral metastasis is higher than in the case of bone metastasis. Additionally, they observed that patients with over 5 CTCs or CTC doublets/clusters were prone to develop more visceral metastases [62]. Also, some studies have reported that the number of CTCs in the peripheral blood of patients with lymph node and visceral metastases was significantly higher than that in patients with bone metastasis or without metastasis [63], [64], [65].
We have developed a device that detects CTCs from breast cancer patient samples in a label-free manner, due to the distinct difference in WBC and CTC size and deformability. The device can be extended to the isolation of CTCs of other cancers and for separation of other sources of liquid biopsy, as long as the targets are larger than the blood cells. We envision that our system could be a promising tool in cancer patient management both in diagnosis and treatment. In metastatic patients, CTC counting could be helpful as an alternative to more invasive methods of diagnosis. It may also provide important prognostic information and could be used in monitoring the treatment efficacy.
There are other commercial microfiltration systems like ScreenCell (ScreenCell) [66] and CellSieveTM (Creatv MicroTech) [67] that provide greater throughput (>5 ml/min) than the MPA-chip (0.5 ml/h). But, filter clogging, damage to the cells, and low CTC capture efficiency and recovery are major disadvantages of these mechanical filtration devices due to their high flow rate and filtration pressure [68], [69], [70].
Additionally, the chip size of the MPA-chip is smaller (5.5 mm × 4 mm) than most systems previously reported, including the commercially available Parsortix (70 mm × 20 mm) [71]. Warkiani and colleagues developed a curvilinear microchannel chip, similar to the ClearCell FX system (Clearbridge Biomedics) (30 mm × 30 mm) [72]. Zhou and co-workers [28] developed a microfluidic device consisting of diamond shape pillars (30 mm × 20 mm), Chen et al. [73] introduced a microfluidic device with micro-ellipse pillars (30 mm × 20 mm), CROSS chip [74] with circular micropillars (40 mm × 40 mm) and other studies (40 mm × 30 mm) [75], (34 mm × 47 mm) [76]. The small size of the MPA-chip resulted in higher throughput of the microfabrication process, since 135 chips can be produced on a single silicon wafer, and 20 chips can be bonded to a standard microscope slide. Comparatively, the other systems mentioned above contain 20 chips on a silicon wafer and finally bond 2 chips to a microscope slide. This feature reduces the time of chip fabrication, and results in saving materials needed to fabricate the chips. Because of the small form factor, the MPA-chip has the potential to decrease the amount of reagents necessary to identify cells (anti-bodies, staining materials, and culture media), and to minimize the risk to lose the rare cells and end up with lower cell viability during post-processing (due to release of CTCs from the chip). It has previously been reported that<50 % of trapped cells are released from inside the chip [24]. Taken together, smaller microfluidic chips such as the MPA-chip show significant advantages such as less sample and reagent demand, short analysis and process time, and precise operation [77], [78], [79], [80].
Importantly, this size minimization facilitates the visualization of the entire separation region of the MPA-chip directly under a 10X lens in the microscope, avoiding movement of the lens or chip, and the tedious bottlenecks of sample post-processing required by numerous CTC isolation techniques [80]. Notably, enumeration, staining, and washing are integrated into MPA-chips.
Furthermore, due to the simplicity of the setup and protocol used, our system can be implemented in a straightforward manner in any research or pathology lab without requiring the use of special equipment, and only using an ordinary syringe pump for CTC isolation. Using our optimized MPA-chip, fabrication, isolation, detection, and even harvesting can be conducted inside the microchip with reduced time and as a result at lower cost.
Conclusion
We introduced a novel microfluidic micro-pillar array chip (MPA-chip) with optimized micro-pillar geometries for separating CTCs from a blood sample based on the size of CTCs and the deformation of leukocytes (WBCs and RBCs). Our results (simulation and experiment) demonstrate that the lozenge geometry have better CTC isolation performance than other micropillar geometries. We found that the cell viability depends mainly on micropillars' geometry, and the lozenge geometry is safer for cells. For the lozenge MPA chip, the capture efficiency is larger than 85 %, the purity is larger than 90 %, and the viability of isolated cells is as high as 97 %, There were no significant differences in the results of capture efficiency, cell viability, and enrichment factor between PC3 and MDA-MB-231 cell lines across all the micro-pillar geometries. Accordingly, our MPA-chip only separates CTCs according to their size and without considering the type of cancer. Our device was also validated by the detection of CTCs in the blood of breast cancer patients. The CTC count showed good potential in monitoring treatment and guiding future individualized treatment. All of the results demonstrate that the lozenge MPA-chips have promising potential for capturing CTCs of breast cancer patients. In this study, we have focused on the influence of the pillar shape and on the validation with patient samples towards clinical relevance. In future work, the influence of the details of the geometrical design (dimensions, distances, positions) could be further studied to improve the CTC isolation performance even more.
CRediT authorship contribution statement
Mehdi Rahmanian: Conceptualization, Methodology, Writing – original draft. Omid Sartipzadeh Hematabad: Software, Investigation. Esfandyar Askari: Validation, Formal analysis. Farhad Shokati: Software, Formal analysis. Atin Bakhshi: Investigation, Resources. Shiva Moghadam: Writing – original draft, Formal analysis. Asiie Olfatbakhsh: Writing – original draft, Formal analysis. Esmat Al Sadat Hashemi: Writing – original draft, Formal analysis. Mohammad Khorsand Ahmadi: Data curation. Seyed Morteza Naghib: Supervision. Nidhi Sinha: Visualization. Jurjen Tel: Visualization. Hossein Eslami Amirabadi: Writing – original draft, Supervision. Jaap M.J. den Toonder: Project administration, Funding acquisition, Supervision. Keivan Majidzadeh-A: Conceptualization, Project administration, Funding acquisition, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.08.005.
Contributor Information
Jaap M.J. den Toonder, Email: J.M.J.d.Toonder@tue.nl.
Keivan Majidzadeh-A, Email: kmajidzadeh@razi.tums.ac.ir.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Balakrishnan S.G., Ahmad M.R., Koloor S.S.R., Petrů M. Separation of ctDNA by superparamagnetic bead particles in microfluidic platform for early cancer detection. J Adv Res. 2021;33:109–116. doi: 10.1016/j.jare.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parada N., Romero-Trujillo A., Georges N., Alcayaga-Miranda F. Camouflage strategies for therapeutic exosomes evasion from phagocytosis. J Adv Res. 2021;31:61–74. doi: 10.1016/j.jare.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saminathan M., Singh K.P., Maity M., Vineetha S., Manjunathareddy G.B., Dhama K., et al. Pathological and immunological characterization of bluetongue virus serotype 1 infection in type I interferons blocked immunocompetent adult mice. J Adv Res. 2021;31:137–153. doi: 10.1016/j.jare.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang F., Li Z., Yu J., Long Y., Zhao Q., Ding X., et al. MicroRNAs secreted by human embryos could be potential biomarkers for clinical outcomes of assisted reproductive technology. J Adv Res. 2021;31:25–34. doi: 10.1016/j.jare.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van de Stolpe A., Den Toonder J.M.J. Circulating tumor cells: what is in it for the patient? A vision towards the future. Cancers. 2014;6 doi: 10.3390/cancers6021195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang S., Wang Y., Onck P., den Toonder J. A concise review of microfluidic particle manipulation methods. Microfluid Nanofluid. 2020;24:24. [Google Scholar]
- 7.den Toonder J. Circulating tumor cells: the Grand Challenge. Lab Chip. 2011;11:375–377. doi: 10.1039/c0lc90100h. [DOI] [PubMed] [Google Scholar]
- 8.Schwarzenbach H. Circulating nucleic acids as biomarkers in breast cancer. Breast Cancer Res. 2013;15:1–9. doi: 10.1186/bcr3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernández M.J.S., Merino J.C.Á., Zubiaurre I.M., García A.F., Rovira P.S., Acosta J.A.L. Clinical relevance associated to the analysis of circulating tumour cells in patients with solid tumours. Clin Transl Oncol. 2009;11:659–668. doi: 10.1007/s12094-009-0421-z. [DOI] [PubMed] [Google Scholar]
- 10.Yap T.A., Lorente D., Omlin A., Olmos D., De Bono J.S. Circulating tumor cells: a multifunctional biomarker. Clin Cancer Res. 2014;20:2553–2568. doi: 10.1158/1078-0432.CCR-13-2664. [DOI] [PubMed] [Google Scholar]
- 11.Rahimzadeh Z., Naghib S.M., Askari E., Molaabasi F., Sadr A., Zare Y., et al. A rapid nanobiosensing platform based on herceptin-conjugated graphene for ultrasensitive detection of circulating tumor cells in early breast cancer. Nanotechnol Rev. 2021;10:744–753. [Google Scholar]
- 12.A. van de Stolpe, K. Pantel, S. Sleijfer, L.W. Terstappen, J.M.J. Den Toonder, Circulating tumor cell isolation and diagnostics: toward routine clinical use, AACR, 2011. [DOI] [PubMed]
- 13.Sadeghi M., Kashanian S., Naghib S.M., Arkan E. A high-performance electrochemical aptasensor based on graphene-decorated rhodium nanoparticles to detect HER2-ECD oncomarker in liquid biopsy. Sci Rep. 2022;12:3299. doi: 10.1038/s41598-022-07230-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.den Toonder J. Circulating tumor cells: the Grand Challenge. Lab Chip. 2011;11(3):375. doi: 10.1039/c0lc90100h. [DOI] [PubMed] [Google Scholar]
- 15.Nagrath S., Sequist L.V., Maheswaran S., Bell D.W., Irimia D., Ulkus L., et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seyfoori A., Seyyed Ebrahimi S.A., Samiei E., Akbari M. Multifunctional hybrid magnetic microgel synthesis for immune-based isolation and post-isolation culture of tumor cells. ACS Appl Mater Interfaces. 2019;11(28):24945–24958. doi: 10.1021/acsami.9b02959. [DOI] [PubMed] [Google Scholar]
- 17.Yu D., Tang L., Dong Z., Loftis K.A., Ding Z., Cheng J., et al. Effective reduction of non-specific binding of blood cells in a microfluidic chip for isolation of rare cancer cells. Biomater Sci. 2018;6(11):2871–2880. doi: 10.1039/c8bm00864g. [DOI] [PubMed] [Google Scholar]
- 18.Spizzo G., Fong D., Wurm M., Ensinger C., Obrist P., Hofer C., et al. EpCAM expression in primary tumour tissues and metastases: an immunohistochemical analysis. J Clin Pathol. 2011;64(5):415–420. doi: 10.1136/jcp.2011.090274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ignatiadis M., Dawson S.-J. Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Ann Oncol. 2014;25(12):2304–2313. doi: 10.1093/annonc/mdu480. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y., Zhao W., Cheng R., Puig A., Hodgson J., Egan M., et al. Label-free inertial-ferrohydrodynamic cell separation with high throughput and resolution. Lab Chip. 2021;21(14):2738–2750. doi: 10.1039/d1lc00282a. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y., Zhao W., Cheng R., Harris B.N., Murrow J.R., Hodgson J., et al. Fundamentals of integrated ferrohydrodynamic cell separation in circulating tumor cell isolation. Lab Chip. 2021;21(9):1706–1723. doi: 10.1039/d1lc00119a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li P., Mao Z., Peng Z., Zhou L., Chen Y., Huang P.-H., et al. Acoustic separation of circulating tumor cells. Proc Natl Acad Sci. 2015;112(16):4970–4975. doi: 10.1073/pnas.1504484112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo L., He Y. Magnetically driven microfluidics for isolation of circulating tumor cells. Cancer Medicine. 2020;9(12):4207–4231. doi: 10.1002/cam4.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarioglu A.F., Aceto N., Kojic N., Donaldson M.C., Zeinali M., Hamza B., et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat Methods. 2015;12(7):685–691. doi: 10.1038/nmeth.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang L.R., Cox E.C., Austin R.H., Sturm J.C. Continuous particle separation through deterministic lateral displacement. Science. 2004;304(5673):987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 26.Salafi T., Zhang Y., Zhang Y. A review on deterministic lateral displacement for particle separation and detection. Nano-Micro Letters. 2019;11:1–33. doi: 10.1007/s40820-019-0308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeming K.K., Ranjan S., Zhang Y. Rotational separation of non-spherical bioparticles using I-shaped pillar arrays in a microfluidic device. Nat Commun. 2013;4:1–8. doi: 10.1038/ncomms2653. [DOI] [PubMed] [Google Scholar]
- 28.Zhou J., Tu C., Liang Y., Huang B., Fang Y., Liang X., et al. The label-free separation and culture of tumor cells in a microfluidic biochip. Analyst. 2020;145(5):1706–1715. doi: 10.1039/c9an02092f. [DOI] [PubMed] [Google Scholar]
- 29.Nasiri R., Shamloo A., Ahadian S., Amirifar L., Akbari J., Goudie M.J., et al. Microfluidic-based approaches in targeted cell/particle separation based on physical properties: fundamentals and applications. Small. 2020;16(29):2000171. doi: 10.1002/smll.202000171. https://doi.org/10.1002/smll.v16.2910.1002/smll.202000171. [DOI] [PubMed] [Google Scholar]
- 30.Shamloo A., Yazdani A., Saghafifar F. Investigation of a two-step device implementing magnetophoresis and dielectrophoresis for separation of circulating tumor cells from blood cells. Eng Life Sci. 2020;20(7):296–304. doi: 10.1002/elsc.202000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khetani S., Mohammadi M., Nezhad A.S. Filter-based isolation, enrichment, and characterization of circulating tumor cells. Biotechnol Bioeng. 2018;115(10):2504–2529. doi: 10.1002/bit.26787. [DOI] [PubMed] [Google Scholar]
- 32.Song Y., Tian T., Shi Y., Liu W., Zou Y., Khajvand T., et al. Enrichment and single-cell analysis of circulating tumor cells. Chem Sci. 2017;8(3):1736–1751. doi: 10.1039/c6sc04671a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fontanella R.A., Sideri S., Di Stefano C., Catizone A., Di Agostino S., Angelini D.F., et al. CD44v8-10 is a marker for malignant traits and a potential driver of bone metastasis in a subpopulation of prostate cancer cells. Cancer Biol Med. 2021;18:788. doi: 10.20892/j.issn.2095-3941.2020.0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang B., Zhang S., Meng J., Min L.i., Luo J., Zhu Z., et al. Evaporation-Induced rGO Coatings for Highly Sensitive and Non-Invasive Diagnosis of Prostate Cancer in the PSA Gray Zone. Adv Mater. 2021;33(40):2103999. doi: 10.1002/adma.202103999. https://doi.org/10.1002/adma.v33.4010.1002/adma.202103999. [DOI] [PubMed] [Google Scholar]
- 35.Eltayeb N.M., Al-Amin M., Yousif A.M., Balakrishnan V., Salhimi S.M. Catharanthus roseus L. extract downregulates the expression profile of motility-related genes in highly invasive human breast cancer cell line MDA-MB-231. Biologia. 2021;76(3):1017–1032. [Google Scholar]
- 36.Simu S., Marcovici I., Dobrescu A., Malita D., Dehelean C.A., Coricovac D., et al. Insights into the Behavior of Triple-Negative MDA-MB-231 Breast Carcinoma Cells Following the Treatment with 17β-Ethinylestradiol and Levonorgestrel. Molecules. 2021;26(9):2776. doi: 10.3390/molecules26092776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu X., Zang X., Lv Y. Detection of circulating tumor cells: Advances and critical concerns. Oncol Lett. 2021;21:1–11. doi: 10.3892/ol.2021.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Connolly S., McGourty K., Newport D. The in vitro inertial positions and viability of cells in suspension under different in vivo flow conditions. Sci Rep. 2020;10:1–13. doi: 10.1038/s41598-020-58161-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian F., Cai L., Chang J., Li S., Liu C., Li T., et al. Label-free isolation of rare tumor cells from untreated whole blood by interfacial viscoelastic microfluidics. Lab Chip. 2018;18(22):3436–3445. doi: 10.1039/c8lc00700d. [DOI] [PubMed] [Google Scholar]
- 40.Li T., Li N., Ma Y., Bai Y.-J., Xing C.-M., Gong Y.-K. A blood cell repelling and tumor cell capturing surface for high-purity enrichment of circulating tumor cells. J Mater Chem B. 2019;7(40):6087–6098. doi: 10.1039/c9tb01649j. [DOI] [PubMed] [Google Scholar]
- 41.Kim S.H., Ito H., Kozuka M., Takagi H., Hirai M., Fujii T. Cancer marker-free enrichment and direct mutation detection in rare cancer cells by combining multi-property isolation and microfluidic concentration. Lab Chip. 2019;19:757–766. doi: 10.1039/c8lc00772a. [DOI] [PubMed] [Google Scholar]
- 42.Liebs S., Eder T., Klauschen F., Schütte M., Yaspo M.-L., Keilholz U., et al. Applicability of liquid biopsies to represent the mutational profile of tumor tissue from different cancer entities. Oncogene. 2021;40:5204–5212. doi: 10.1038/s41388-021-01928-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hong B., Zu Y. Detecting circulating tumor cells: current challenges and new trends. Theranostics. 2013;3:377. doi: 10.7150/thno.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan S.J., Lakshmi R.L., Chen P., Lim W.-T., Yobas L., Lim C.T. Versatile label free biochip for the detection of circulating tumor cells from peripheral blood in cancer patients. Biosens Bioelectron. 2010;26:1701–1705. doi: 10.1016/j.bios.2010.07.054. [DOI] [PubMed] [Google Scholar]
- 45.Nasiri R., Shamloo A., Akbari J., Tebon P., Dokmeci M.R., Ahadian S. Design and simulation of an integrated centrifugal microfluidic device for CTCs separation and cell lysis. Micromachines. 2020;11:699. doi: 10.3390/mi11070699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shamloo A., Vatankhah P., Bijarchi M.A. Numerical optimization and inverse study of a microfluidic device for blood plasma separation. Eur J Mech-B/Fluids. 2016;57:31–39. [Google Scholar]
- 47.Krog B.L., Henry M.D. Biomechanics of the circulating tumor cell microenvironment. Biomech Oncol. 2018:209–233. doi: 10.1007/978-3-319-95294-9_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marrella A., Fedi A., Varani G., Vaccari I., Fato M., Firpo G., et al. High blood flow shear stress values are associated with circulating tumor cells cluster disaggregation in a multi-channel microfluidic device. PLoS ONE. 2021;16(1):e0245536. doi: 10.1371/journal.pone.0245536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paizal J.P., Au S.H., Bakal C. Squeezing through the microcirculation: survival adaptations of circulating tumour cells to seed metastasis. Br J Cancer. 2021;124:58–65. doi: 10.1038/s41416-020-01176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cognart H.A., Viovy J.-L., Villard C. Fluid shear stress coupled with narrow constrictions induce cell type-dependent morphological and molecular changes in SK-BR-3 and MDA-MB-231 cells. Sci Rep. 2020;10:1–14. doi: 10.1038/s41598-020-63316-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hyler A.R., Baudoin N.C., Brown M.S., Stremler M.A., Cimini D., Davalos R.V., et al. Fluid shear stress impacts ovarian cancer cell viability, subcellular organization, and promotes genomic instability. PLoS ONE. 2018;13(3):e0194170. doi: 10.1371/journal.pone.0194170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Z., Huang F., Du J., Shu W., Feng H., Xu X., et al. Rapid isolation of cancer cells using microfluidic deterministic lateral displacement structure. Biomicrofluidics. 2013;7(1):011801. doi: 10.1063/1.4774308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loutherback K., Chou K.S., Newman J., Puchalla J., Austin R.H., Sturm J.C. Improved performance of deterministic lateral displacement arrays with triangular posts. Microfluid Nanofluid. 2010;9(6):1143–1149. [Google Scholar]
- 54.Xu L., Mao X., Imrali A., Syed F., Mutsvangwa K., Berney D., et al. Optimization and evaluation of a novel size based circulating tumor cell isolation system. PLoS ONE. 2015;10(9):e0138032. doi: 10.1371/journal.pone.0138032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Renier C., Pao E., Che J., Liu H.E., Lemaire C.A., Matsumoto M., et al. Label-free isolation of prostate circulating tumor cells using Vortex microfluidic technology. npj Precis Oncol. 2017;1(1) doi: 10.1038/s41698-017-0015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sollier E., Go D.E., Che J., Gossett D.R., O'Byrne S., Weaver W.M., et al. Size-selective collection of circulating tumor cells using Vortex technology. Lab Chip. 2014;14:63–77. doi: 10.1039/c3lc50689d. [DOI] [PubMed] [Google Scholar]
- 57.Che J., Yu V., Dhar M., Renier C., Matsumoto M., Heirich K., et al. Classification of large circulating tumor cells isolated with ultra-high throughput microfluidic Vortex technology. Oncotarget. 2016;7:12748. doi: 10.18632/oncotarget.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riahi R., Gogoi P., Sepehri S., Zhou Y., Handique K., Godsey J., et al. A novel microchannel-based device to capture and analyze circulating tumor cells (CTCs) of breast cancer. Int J Oncol. 2014;44:1870–1878. doi: 10.3892/ijo.2014.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hvichia G.E., Parveen Z., Wagner C., Janning M., Quidde J., Stein A., et al. A novel microfluidic platform for size and deformability based separation and the subsequent molecular characterization of viable circulating tumor cells. Int J Cancer. 2016;138:2894–2904. doi: 10.1002/ijc.30007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen J., Ye C., Dong J., Cao S., Hu Y., Situ B., et al. Metabolic classification of circulating tumor cells as a biomarker for metastasis and prognosis in breast cancer. J Translational Med. 2020;18:1–14. doi: 10.1186/s12967-020-02237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cristofanilli M., Pierga J.-Y., Reuben J., Rademaker A., Davis A.A., Peeters D.J., et al. The clinical use of circulating tumor cells (CTCs) enumeration for staging of metastatic breast cancer (MBC): international expert consensus paper. Critical Rev Oncol/Hematol. 2019;134:39–45. doi: 10.1016/j.critrevonc.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 62.Yu T., Wang C., Xie M., Zhu C., Shu Y., Tang J., et al. Heterogeneity of CTC contributes to the organotropism of breast cancer. Biomed Pharmacother. 2021;137 doi: 10.1016/j.biopha.2021.111314. [DOI] [PubMed] [Google Scholar]
- 63.Zhang X., Lu X., Gao W., Wang Y., Jia C., Cong H. A label-free microfluidic chip for the highly selective isolation of single and cluster CTCs from breast cancer patients. Transl Oncol. 2021;14 doi: 10.1016/j.tranon.2020.100959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pantano F., Rossi E., Iuliani M., Facchinetti A., Simonetti S., Ribelli G., et al. Dynamic changes of receptor activator of nuclear factor-κB expression in circulating tumor cells during denosumab predict treatment effectiveness in metastatic breast cancer. Sci Rep. 2020;10:1–12. doi: 10.1038/s41598-020-58339-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu D., Wu J., Lin C., Andriani L., Ding S., Shen K., et al. Breast subtypes and prognosis of breast cancer patients with initial bone metastasis: a population-based study. Front Oncol. 2020;10:2699. doi: 10.3389/fonc.2020.580112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Desitter I., Guerrouahen B.S., Benali-Furet N., Wechsler J., Jänne P.A., Kuang Y., et al. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res. 2011;31:427–441. [PubMed] [Google Scholar]
- 67.Adams D.L., Zhu P., Makarova O.V., Martin S.S., Charpentier M., Chumsri S., et al. The systematic study of circulating tumor cell isolation using lithographic microfilters. RSC Adv. 2014;4(9):4334–4342. doi: 10.1039/C3RA46839A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cho H., Kim J., Song H., Sohn K.Y., Jeon M., Han K.-H. Microfluidic technologies for circulating tumor cell isolation. Analyst. 2018;143(13):2936–2970. doi: 10.1039/c7an01979c. [DOI] [PubMed] [Google Scholar]
- 69.Zhou M.-D., Hao S., Williams A.J., Harouaka R.A., Schrand B., Rawal S., et al. Separable bilayer microfiltration device for viable label-free enrichment of circulating tumour cells. Sci Rep. 2014;4(1) doi: 10.1038/srep07392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng S., Lin H.K., Lu B.o., Williams A., Datar R., Cote R.J., et al. 3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood. Biomed Microdevices. 2011;13(1):203–213. doi: 10.1007/s10544-010-9485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller M.C., Robinson P.S., Wagner C., O'Shannessy D.J. The Parsortix™ cell separation system—A versatile liquid biopsy platform. Cytometry Part A. 2018;93(12):1234–1239. doi: 10.1002/cyto.a.23571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Warkiani M.E., Khoo B.L., Wu L., Tay A.K.P., Bhagat A.A.S., Han J., et al. Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nat Protoc. 2016;11(1):134–148. doi: 10.1038/nprot.2016.003. [DOI] [PubMed] [Google Scholar]
- 73.Chen H., Cao B., Sun B., Cao Y., Yang K., Lin Y.-S. Highly-sensitive capture of circulating tumor cells using micro-ellipse filters. Sci Rep. 2017;7:1–10. doi: 10.1038/s41598-017-00232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ribeiro-Samy S., Oliveira M.I., Pereira-Veiga T., Muinelo-Romay L., Carvalho S., Gaspar J., et al. Fast and efficient microfluidic cell filter for isolation of circulating tumor cells from unprocessed whole blood of colorectal cancer patients. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-44401-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Armbrecht L., Rutschmann O., Szczerba B.M., Nikoloff J., Aceto N., Dittrich P.S. Quantification of protein secretion from circulating tumor cells in microfluidic chambers. Adv Sci. 2020;7(11):1903237. doi: 10.1002/advs.201903237. https://doi.org/10.1002/advs.v7.1110.1002/advs.201903237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kang Y.T., Hadlock T., Lo T.W., Purcell E., Mutukuri A., Fouladdel S., et al. Dual-Isolation and profiling of circulating tumor cells and cancer exosomes from blood samples with melanoma using immunoaffinity-based microfluidic interfaces. Adv Sci. 2020;7:2001581. doi: 10.1002/advs.202001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maertens Y., Humberg V., Erlmeier F., Steffens S., Steinestel J., Bögemann M., et al. Comparison of isolation platforms for detection of circulating renal cell carcinoma cells. Oncotarget. 2017;8:87710. doi: 10.18632/oncotarget.21197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Papadaki M.A., Sotiriou A.I., Vasilopoulou C., Filika M., Aggouraki D., Tsoulfas P.G., et al. Optimization of the enrichment of circulating tumor cells for downstream phenotypic analysis in patients with non-small cell lung cancer treated with anti-pd-1 immunotherapy. Cancers. 2020;12:1556. doi: 10.3390/cancers12061556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bankó P., Lee S.Y., Nagygyörgy V., Zrínyi M., Chae C.H., Cho D.H., et al. Technologies for circulating tumor cell separation from whole blood. J Hematol Oncol. 2019;12:1–20. doi: 10.1186/s13045-019-0735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jackson J.M., Witek M.A., Kamande J.W., Soper S.A. Materials and microfluidics: enabling the efficient isolation and analysis of circulating tumour cells. Chem Soc Rev. 2017;46:4245–4280. doi: 10.1039/c7cs00016b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.