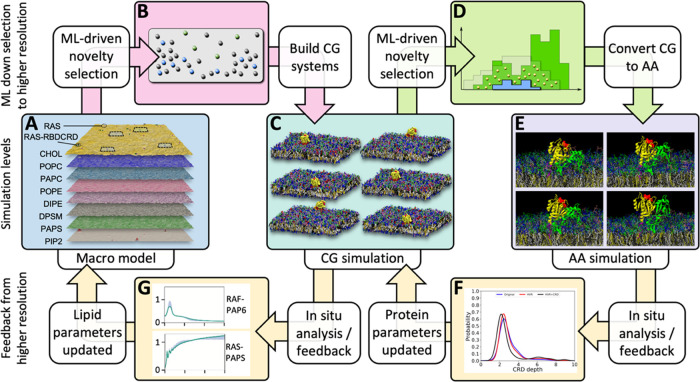
Figure 1.
Three-scale MuMMI scheme. As the continuum macro model (A) evolves over time, ML-driven sampling is used to identify a diverse set of configurations (B) from the continuum model to spawn CG simulations (C) in a DL-based latent space. From the resulting CG simulations, candidate CG snapshots to convert to AA simulations are chosen in a three-dimensional parameter space (D). The subsequent AA simulations (E) are analyzed in real time to provide updated protein parameters (F) to the CG simulations. Concurrently, in situ analysis of the CG simulations refines the protein–lipid interactions (G) for the macro model.