Figure 4.
6mA is related to alternative splicing
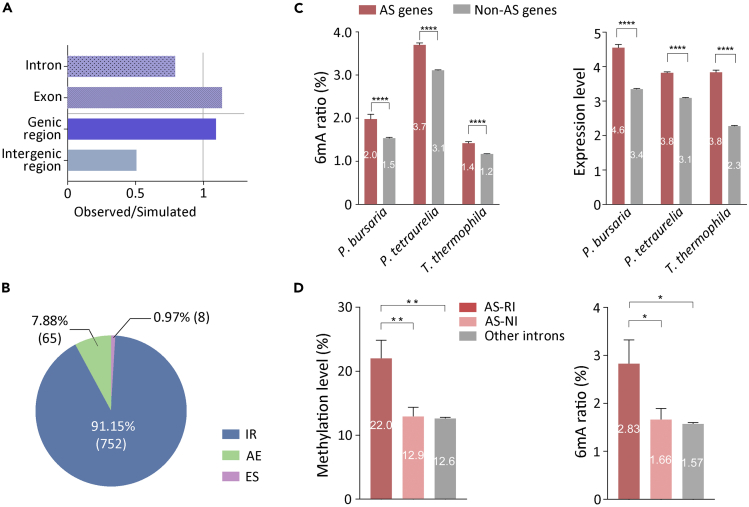
(A) Comparison of observed versus simulated distributions of 6mA in P. bursaria genome suggests that 6mA is enriched in exons rather than introns.
(B) The ratio of different alternative splicing events (IR: intron retention, AE: alternative 5' or 3′ splice-site selection, ES: exon skipping) in P. bursaria.
(C) Statistical analysis of 6mA ratio (left panel) and expression level (right panel) indicated by log2 transformation of FPKM (fragments per kilobase per million) in P. bursaria (n1 = 360; n2 = 17,442), P. tetraurelia (n1 = 3369; n2 = 30,048), and T. thermophila (n1 = 1,066; n2 = 25,151) shows that AS genes contain obviously higher expression levels and amounts of 6mA compared with other genes. Student’s t test was performed, and n represents the number of corresponding genes per group. Data are represented as mean ± SEM. ∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05; ns: not significant, p > 0.05. 6mA ratio was defined as the number of methylated adenine sites divided by the total adenine sites (6mA/A) in the corresponding genomic sequence.
(D) Statistical analysis of methylation level (left panel) and 6mA ratio (right panel) in P. bursaria demonstrates that the level of 6mA in retained introns of AS (AS-RI) genes (n = 150) is higher than that in normal introns of AS (AS-NI) genes (n = 423) and other introns of non-AS (other introns) genes (n = 24,818). Student’s t test was performed, and n represents the number of corresponding introns per group. Data are represented as mean ± SEM. ∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05; ns: not significant (p > 0.05). Methylation level here was calculated as average methylation level of per intron.