Extended Figure 8. Two biological replicates of Hi-C analysis of G1-sorted nuclei with RAD21 cleaved in NBS1 (with Supplementary Fig. 3).
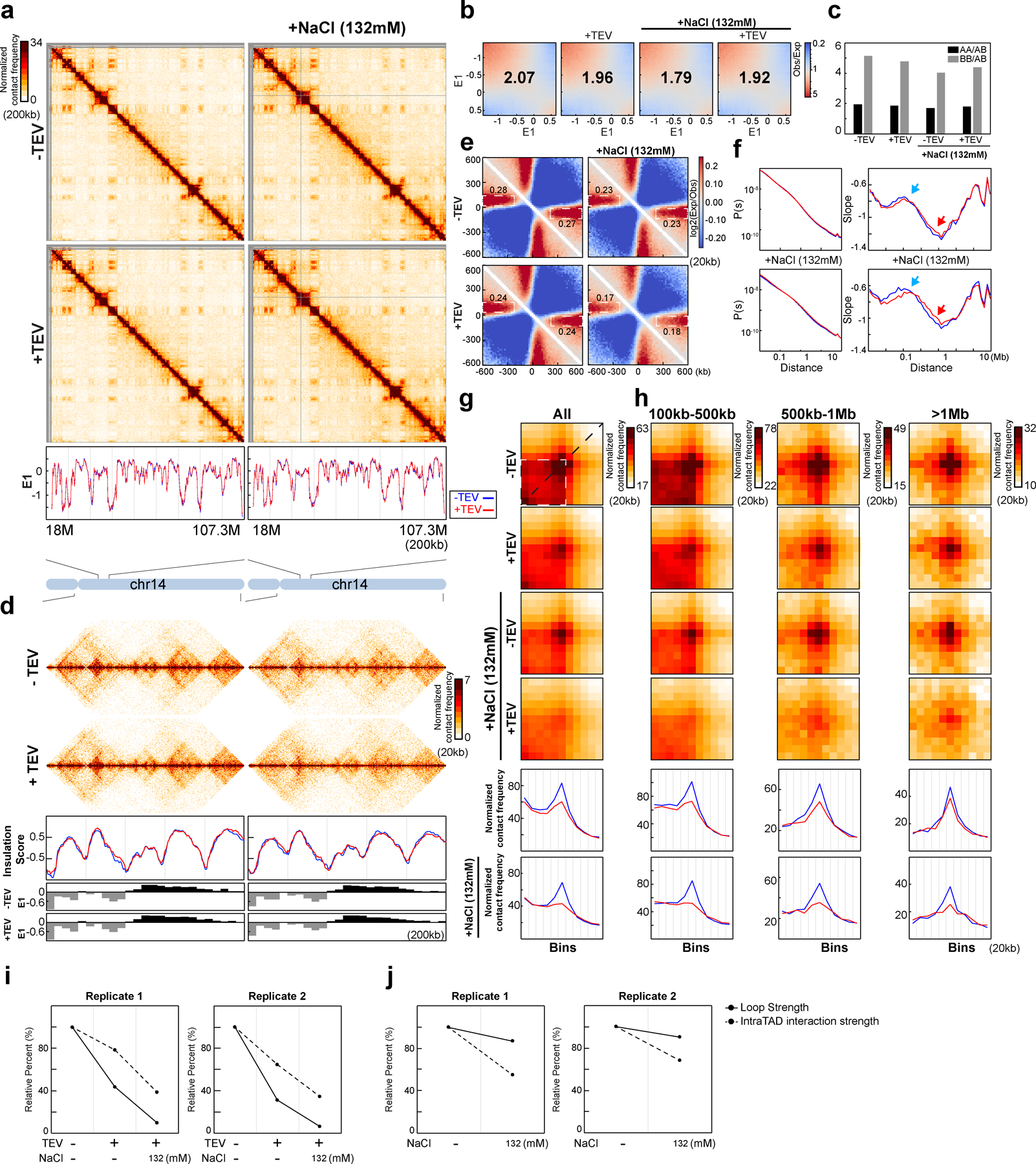
(a) Hi-C maps for G1-sorted HAP1-RAD21TEV nuclei treated with TEV in specified buffers as shown. Data for the 18–107.3 Mb region of chromosome 14 is shown. Bottom, E1 cross the same region. (b) Saddle plots for G1-sorted HAP1-RAD21TEV nuclei treated with TEV in specified buffer as shown. Numbers indicate compartment strength. (c) Interaction strength of compartments. Bars represent strength of compartment interactions for each sample as described in Fig. 1h. (d) Hi-C interaction maps for G1-sorted HAP1-RAD21TEV nuclei treated with TEV in specified buffers as shown. Data for the 29–34 Mb region of chromosome 14 is shown. Middle panels indicate insulation profiles for the 29–34 Mb regions of chromosome 14. Blue and red lines represent without and with TEV protease treatment, respectively. The lower panels indicate compartment E1 across the region. (e) Aggregate Hi-C data at TAD boundaries identified in each condition as shown. Numbers at the sides of the cross indicate strength of boundary-anchored stripes using the mean values of interaction frequency within the white dashed boxes. (f) P(s) plots (left panels), and derivatives of P(s) plots (right panels) for Hi-C data from nuclei treated with TEV as shown. Blue arrows indicate the signature of cohesin loops in each condition. Red arrows indicate changes of contact frequency at 2Mb. (g) Aggregated Hi-C data at loops as in Fig. 2j. Lower panel: average Hi-C signals along the blue dashed line shown in the left Hi-C panel. (h) Aggregated Hi-C data at chromatin loops of three different loop sizes as shown. Lower panels: average Hi-C signals along the blue dashed line shown in the left Hi-C map in Fig. 2j. (i) Quantification of loop strength and intra-TAD interaction strength obtained with G1-sorted HAP1-RAD21TEV nuclei treated with TEV in specified buffer as shown (two biological replicates). (j) Quantification of loop strength and intra-TAD interaction strength in each condition as shown. Loop strength and intra-TAD interaction strength were normalized as in Fig. 3i. See source data for numerical data.
