Extended Figure 4. Cleaving RAD21 in NBS1 dissociates and releases cohesin components.
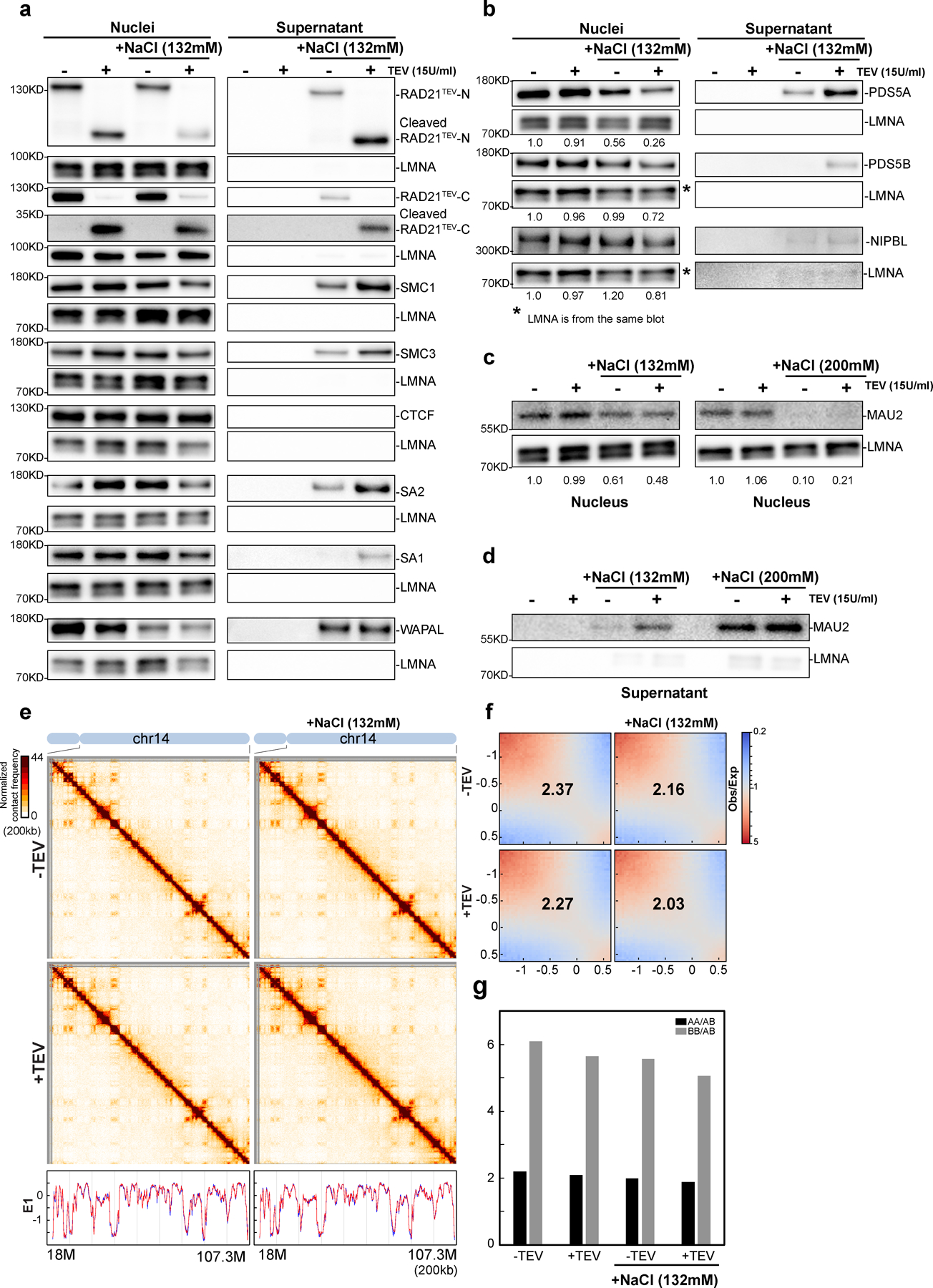
Western blot analysis of nuclear retention of cohesin in RAD21TEV nuclei treated with TEV in the specified buffer as shown. Left panels indicated as nuclei show cohesin components in nuclei detected using antibodies as described in the right panels of a-c. Right panels show supernatant of released cohesin components. Cohesin components from supernatants were separated and detected using the same antibodies as used for the Western blots shown on the right panels of a-c. For all western blot analyses, LMNA was used as the loading controls. (a) A biological replicate of cohesin subunits retained in nuclei or released to the supernatant treated with TEV in specified buffer as shown. The antibodies used here are the same as Fig. 3a. (b) Western blot analysis of PDS5A, PDS5B and NIPBL retained in nuclei or released to the supernatant treated with TEV in specified buffer as shown. (c) and (d) Western analysis of MAU2 (SCC4) retained in nuclei (c) and released to the supernatant (d) treated with TEV in the specified buffer as shown. Cleaving RAD21 in NBS1 has little effects on compartmentalization and CTCF binding (e) Hi-C interaction maps for HAP1-RAD21TEV nuclei treated with TEV as shown. Data for the 18–107.3 Mb region of chromosome 14 is shown. Bottom, eigenvector E1 profiles across the same region. (f) Saddle plots for HAP1-RAD21TEV nuclei treated with TEV as shown. The numbers indicate compartment strength. (g) Interaction strength of compartments. The bars represent the strength of compartment interactions for each sample as described in Fig1h. Source numerical data and unprocessed blots are available in source data.
