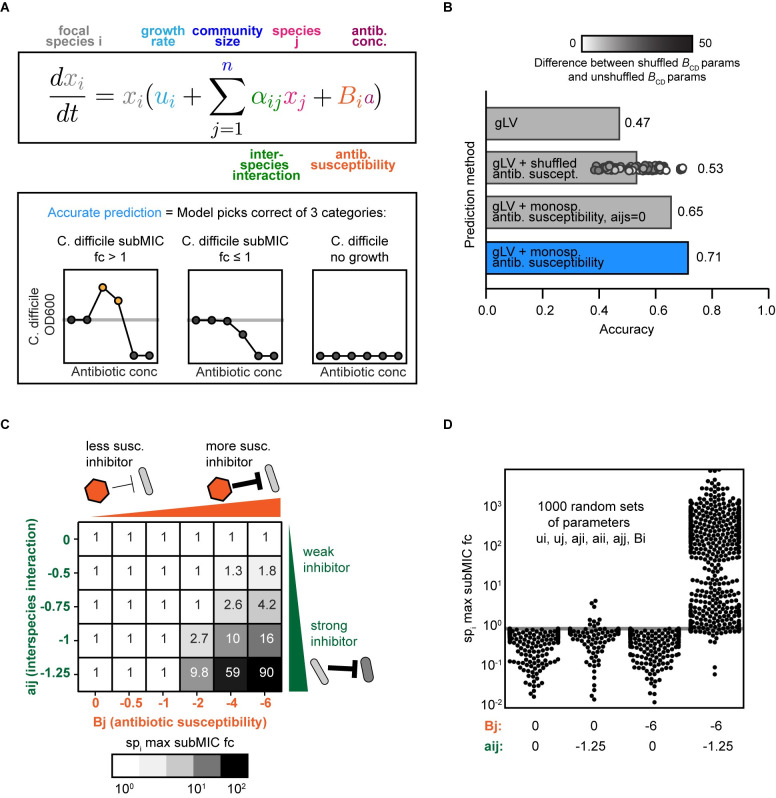
Fig 4. A modified generalized Lotka–Volterra model of community dynamics in response to antibiotics captures trends in antibiotic concentrations lower than the MIC in the presence of antibiotic sensitive biotic inhibitors.
(A) Top: Schematic of the antibiotic expansion of the gLV model. Bottom: Schematic of accuracy metric used in panel B. (B) Qualitative accuracy of multiple models for pairwise and multispecies communities. Models include (1) standard gLV model lacking an antibiotic term; (2) gLV model with randomly shuffled antibiotic susceptibility parameters (average of 100 permutations), and 100 shuffled values shown as points, shaded according to the absolute value of the difference between the shuffled C. difficile antibiotic susceptibility parameter and the original C. difficile antibiotic susceptibility parameter; (3) gLV model with antibiotic susceptibility terms inferred from monospecies with all interaction parameters set to zero (aij = 0, where i! = j); (4) full gLV model with antibiotic susceptibility terms inferred from monospecies data. (C) Simulated maximum subMIC fold changes for a focal species in a pairwise community for 48 h for a representative parameter set. Growth rates of the 2 species are equal (ui = uj = 0.25), intraspecies interactions are equal (aii = ajj = −0.8), and initial ODs are equal (xi(0) = xj(0) = 0.0022). The interspecies interaction coefficients aji are set to 0 and Bi is set to −2. (D) Simulated max subMIC fold change for a focal species in a pairwise community for 1,000 randomly sampled parameter sets. Parameters Bj and aij are set to constant values of 0 or −6 and 0 or −1.25, respectively. Other parameters were randomly sampled between upper and lower bounds. The lower and upper bounds on each parameter value are aji (−1.25, 1.25), growth rates (0, 1), intraspecies interactions (−1.25, 0), Bi (−6, 0). Gray horizontal line at y = 1 indicates no change in growth compared to the no antibiotic condition. The data and modeling scripts underlying panels BCD in this figure can be found in DOI: 10.5281/zenodo.7726490. MIC, minimum inhibitory concentration; subMIC, sub-minimum inhibitory concentration.