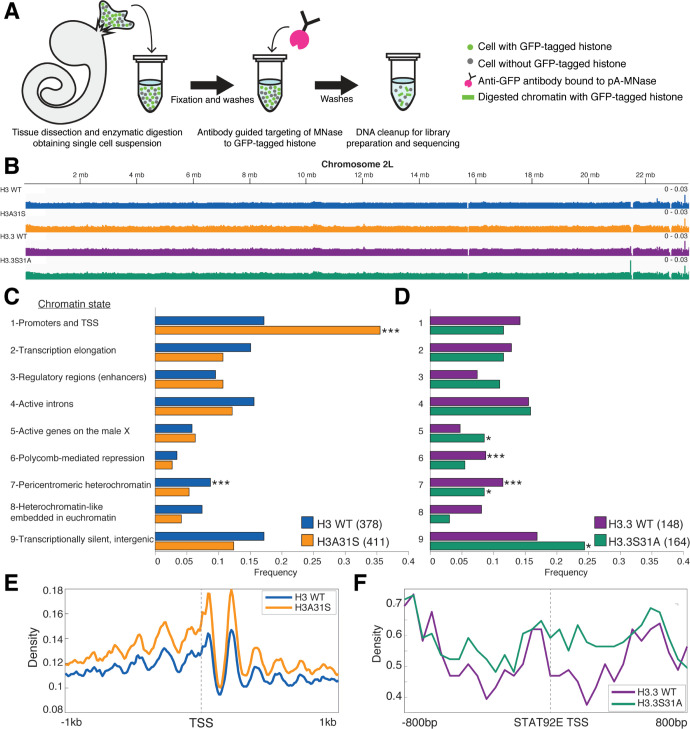
Fig 7. The single residue substitution causes mislocalization of mutant histones to different chromatin contexts.
(A) Germline tumor testes with GSCs expressing GFP-tagged histones were used in a ChIC assay targeting MNase digestion to the tagged histones. The cleaved DNA was then prepared for next-generation DNA sequencing. (B) Genome browser view of Chromosome 2L showing incorporation of transgenic histones genome-wide. (C, D) Genomic enrichment was compared between each WT histone and the mutant counterpart within predefined regions of the specified chromatin states. The frequency of regions in each of the states that showed differential enrichment is indicated. *P < 0.05, ***P < 0.001. The data underlying these panels can be found in S7 Table. (E) Density of reads for H3 WT and H3A31S is plotted +/−1 kb centered on TSSs from chromatin state 1. (E) The density of H3.3 WT and H3.3S31A reads is shown +/− 800 bp centered at the stat92E TSS. ChIC, chromatin immunocleavage; GSC, germline stem cell; TSS, transcription start site; WT, wild-type.