Abstract
Binding kinetics play an important role in cancer diagnosis and therapeutics. However, current methods of quantifying binding kinetics fail to consider the three-dimensional environment that drugs and imaging agents experience in biological tissue. In response, a methodology to assay agent binding and dissociation in 3D tissue culture was developed using paired-agent molecular imaging principles. To test the methodology, the uptakes of ABY-029 (an IRDye 800CW-labeled epidermal growth factor receptor (EGFR)-targeted antibody-mimetic) and IRDye 700DX-carboxylate in 3D spheroids were measured in four different human cancer cell lines throughout staining and rinsing. A compartment model (optimized for the application) was then fit to the kinetic curves of both imaging agents to estimate binding and dissociation rate constants of the EGFR targeted ABY-029 agent. A linear correlation was observed between apparent association rate constant () and the receptor concentration experimentally and in simulations (, ). Additionally, a similar binding affinity profile compared to a gold standard method was determined by this model. This low-cost methodology to quantify imaging agent or drug binding affinity in clinically relevant 3D tumor spheroid models, can be used to guide timing of imaging in molecular guided surgery and could have implications in drug development.
Keywords: Tracer Kinetic Modeling, Drug Affinity, Fluorescence, Epidermal Growth Factor Receptor
1. Introduction
Enormous efforts have been undertaken in drug discovery and development to identify targeted molecules that possess suitable characteristics for cancer targeted diagnostics and therapeutics (1–3). The likelihoods of a drug or imaging agent to bind and dissociate from the biological molecule it is targeted to play important roles in evaluating and optimizing drug/imaging agent dosing, and in the case of imaging agent, timing of imaging after dosing. One aspect of this is binding affinity, , which represents the likelihood a molecule to bind to the targeted site divided by the likelihood it will decouple from the binding site (4). Binding affinity is often reported by , the equilibrium dissociation constant, which is the inverse of affinity and has units of concentration. However, there is evidence that the binding affinity may not be sufficient to provide a complete understanding of drug binding efficacy (5;6). Not only does the affinity parameter mask the temporal characteristic of binding (how fast does binding equilibrium occur), the physiological density of the biological molecular site being targeted (the availability of drug or imaging agent targets) affects likelihood of binding and is ignored by the affinity parameter (7–9).
Several techniques are currently used to analyze the binding kinetics of a compound to a target. Surface plasmon resonance (SPR) is a common biosensor-based technique for characterizing the affinity and kinetics of drug binding that is now considered the gold standard for academic and industry applications (10). This technique works by tethering a binding site to a surface and monitoring a change in mass as the drug/agent binds to the affixed target. It is a highly sensitive and precise tool that requires very low sample volumes (11). However, SPR requires immobilization of functional targets that make it less ideal for some applications; for instance, captured antibodies can nonspecifically bind small molecules (12). In addition, this technique can be costly and less accessible, therefore making it less suitable in some research settings. Biolayer interferometry (BLI) is an alternative for evaluating binding kinetics: this is an optical biosensor-based technique that monitors the molecules bound in real time by analyzing a shift of interference pattern of white light reflected from the biosensor tip (13). It is a convenient and reliable method to perform kinetic measurements with relatively high throughput. However, it also requires immobilization of target binding sites, and the detection sensitivity is lower than SPR (14).
The major drawback of target immobilization in SPR and BLI is that it can affect the binding characteristics owing to the restricted movement (15), and make it difficult to assume similarity of measured parameters to drug-receptor interaction in the native environment (i.e., living cells). Additionally, many of the current techniques for cancer diagnostics and therapeutic drug screening rely on 2-D in vitro experiments, where cell lines are cultured in artificial conditions that do not fully recapitulate the tumor microenvironment morphologically and physiologically (16–20). Although preclinical studies rely heavily on animal tumor models, they may introduce a mixture of confounding effects on analyses. These factors may all contribute to the poor success of anticancer drug development—new cancer drug approval rates are <5% (18). In response, we developed a paired-agent fluorescence imaging method for evaluating fluorescently labelled drug or imaging agent binding kinetics in 3-D cancer spheroids. In this study, the goal was to demonstrate this cost-effective, easy-to-adopt method to independently estimate association and disassociation rate constants in a more relevant and translatable in vitro system. One example of a specific application for this assay has been its use in guiding optimal staining and rinsing strategies of excised lymph nodes to aid in rapid assessment of cancer-bearing status for cancer staging (21–25).
2. Methods and Materials
2.1. Compartment models for paired-agent kinetic analysis
Paired-agent modeling encompasses all molecular imaging methods that incorporate coadministration of a targeted imaging agent with a suitable control (untargeted) imaging agent. The signal from the control agent is used to account for nonspecific uptake and delivery effects on the targeted agent that can obfuscate measurement of specific binding. In this work, a methodology was developed to approximate targeted imaging agent/drug binding kinetics, which was derived from relatively simplistic compartment modeling, that has been previously described (26). The specifics of the modeling employed here are depicted in Fig. 1. Briefly, the concentration of the control and targeted agents in the staining solution is represented as and , respectively. Rate constants, and , represent transport of the the agents between the staining solution concentration and the interstitial “free” space compartment , which in this case is the agarose gel where the cancer spheroids are embedded (where for the control agent and for the targeted agent). Assuming first order kinetics, the following differential equation can be constructed to model the rate of change of the control agent in the tissue medium (gel) as a function of time, :
| (1) |
Eq. 1 can also be applied to describe the uptake of targeted agent in the interstitial space in the absence of targeted binding. However, in the presence of targeted binding (cancer in this case) an additional specific binding compartment, represented by the concentration of bound targeted agent, , was added. The first-order system of differential equations modeling the targeted agent was then defined as:
| (2) |
where and represent the rate constants governing the rate of association and dissociation of targeted agent to and from the binding site of the targeted biological molecule, respectively.
Figure 1.
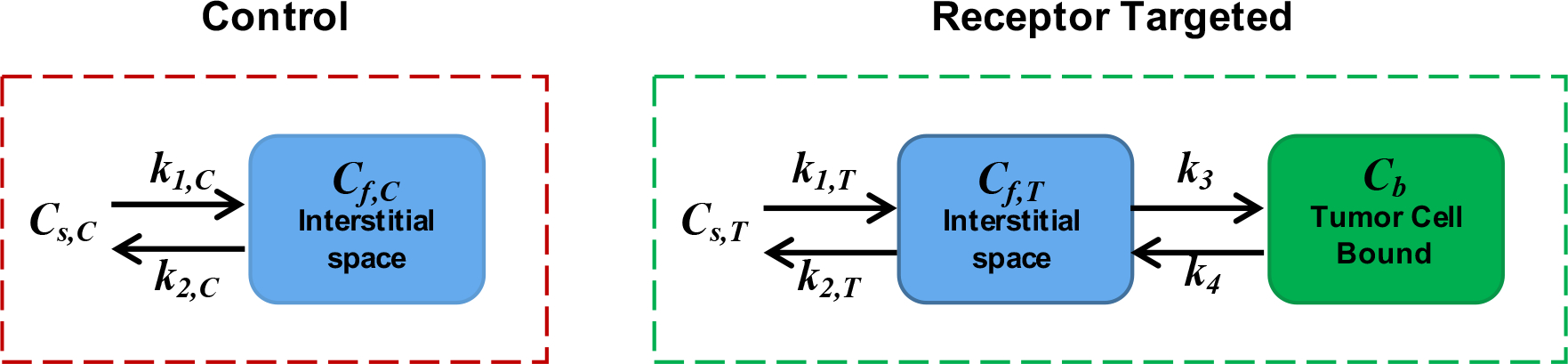
The compartment model for the control and targeted imaging agent. represents the concentration of imaging agent staining solution, represents the concentration of imaging agent in the interstitial free space, represents the concentration of specifically bound targeted agent. and are the rates of exchange of the imaging agent between staining/rinsing solution and the interstitial space. and are the rate constants of agent-to-target-site association and dissociation, respectively.
If the targeted agent concentration is maintained at “trace” levels (i.e., binding sites remain below ~5% bound by the targeted agent), can be considered a constant (27). Otherwise, the binding will be governed by second-order kinetics to incorporate saturation, such that:
| (3) |
where represents the likelihood of binding per concentration of available binding sites, and is total concentration of receptors available to targeted agent prior to binding (28).
2.2. In vitro 3-D cell culture
Four human cancer cell lines, each expressing a different level of epidermal growth factor receptor (EGFR), were selected for this study. A431 (ATCC, Mansassas, VA), a human epidermoid carcinoma known to express a large amount of EGFR (29), was chosen to represent a high EGFR expressing cell line. MDA-MB-231 (ATCC, Mansassas, VA), a human breast adenocarcinoma cell line, and BT-474 (ATCC, Mansassas, VA), a human mammary ductal carcinoma cell line, were selected to represent moderate and low levels of EGFR expression, respectively (30). SW620 (provided by Gibbs Lab at Oregon Health and Science University), a human colon adenocarcinoma cell line, was selected as a negative control owing to its negligible EGFR expression (31). All cell lines were cultured in Dulbecco’s modified Eagle medium (DMEM, Corning, NY) with supplements of 10% fetal bovine serum (FBS, HyClone, Logan, Utah) and 1% Penicillin/Streptomycin (Gibco, Grand Island, NY) at 37 °C in a humidified atmosphere of 5% CO2 in air.
To better recapitulate the in vivo microenvironment morphologically and physiologically (16–20), cancer cell spheroids were cultured to represent tumors in a 3-D architecture. Spheroid formation has been described previously (32). In brief, 1×104 suspended cells in 150 μL of culture medium containing 0.24% (w/v) methycellulose were seeded in each well of a round-bottom, non-tissue treated 96-well plate (Falcon, NY) and incubated for 3 days. Spheroid formation was confirmed under bright field microscopy.
Tumor spheroids were transferred from culture medium to a 1.5-mL microcentrifuge tube via a micropipette. Thirty spheroids were collected in each tube, then all spheroids were allowed to settle to the bottom of the tube by gravity. Medium supernatant was then removed, and spheroids were washed twice by phosphate-buffered saline (PBS), then resuspended in 250 μL of fresh PBS. The resuspended spheroids were mixed with 250 μL of 0.6% (w/v) warm agarose gel solution and dispensed immediately into a 12-well plate. The plate was gently swirled to allow the spheroid-gel solution to cover the entire bottom surface of the plate, and all trapped air to be released. The plate was placed flat on ice for 15 min to allow the agarose solution to cool until it gelled completely. This procedure created an in vitro platform that encapsulated cancer spheroids in 0.3% agarose gel to further perform assessment in a more biologically relevant 3-D environment.
2.3. Imaging protocol
The targeted agent ABY-029, a Good Laboratory Practices (GLP)-produced anti-EGFR Affibody molecule labeled with IRDye-800CW that is in phase 0 clinical trials (33; 34), was paired with IRDye-700DX carboxylate, a non-targeted small molecule imaging agent. The GLP-synthesized ABY-029 stock solutions were provided by the Samkoe lab at Dartmouth College (35). The IRDye-700DX carboxylate was converted from IRDye-700DX NHS ester (Licor Biosciences, Lincoln, NE) by dissolving IRDye-700DX NHS in PBS at a pH of 8.5. The solution was protected from light and gently stirred at room temperature for 5 h.
All images were acquired with an 85-μm resolution planar fluorescence imaging system (Pearl® Impulse, Licor Biosciences, Lincoln, NE) to image white light reflectance and fluorescence emission at 700–740 nm and 800–840 nm from the excitation at 685 nm and 785 nm, respectively. Pre-staining images of agarose gel embedded spheroids were acquired to measure and ultimately remove background signal and autofluorescence. A 5-mL equimolar mixed solution of ABY-029 and IRDye-700DX carboxylate at a trace-level of 2-nM concentration was mixed with 2% (w/v) BSA in PBS and added onto the surface of the gel in each well. Caution was taken to slowly dispense the solution so that there was minimal disruption to the sample. The encapsulated spheroids were stained for 1 h total at room temperature, during which time, the 12-well plate was covered with aluminum foil to protect it from light. During the staining process, staining solution was removed and replaced to allow for images to be acquired on the Pearl imaging system at 15-min intervals to monitor the fluorescence of ABY-029 and IRDye-700DX carboxylate. Excess solution on the gel surface was carefully removed to prevent signal measurement errors caused by staining solution residue. The embedded stained spheroids were then followed by four subsequent rinses in 5 mL of PBS with 15 min per rinse. Images were acquired at each rinse after carefully removing all rinsing saline from the well (Fig. 2).
Figure 2.
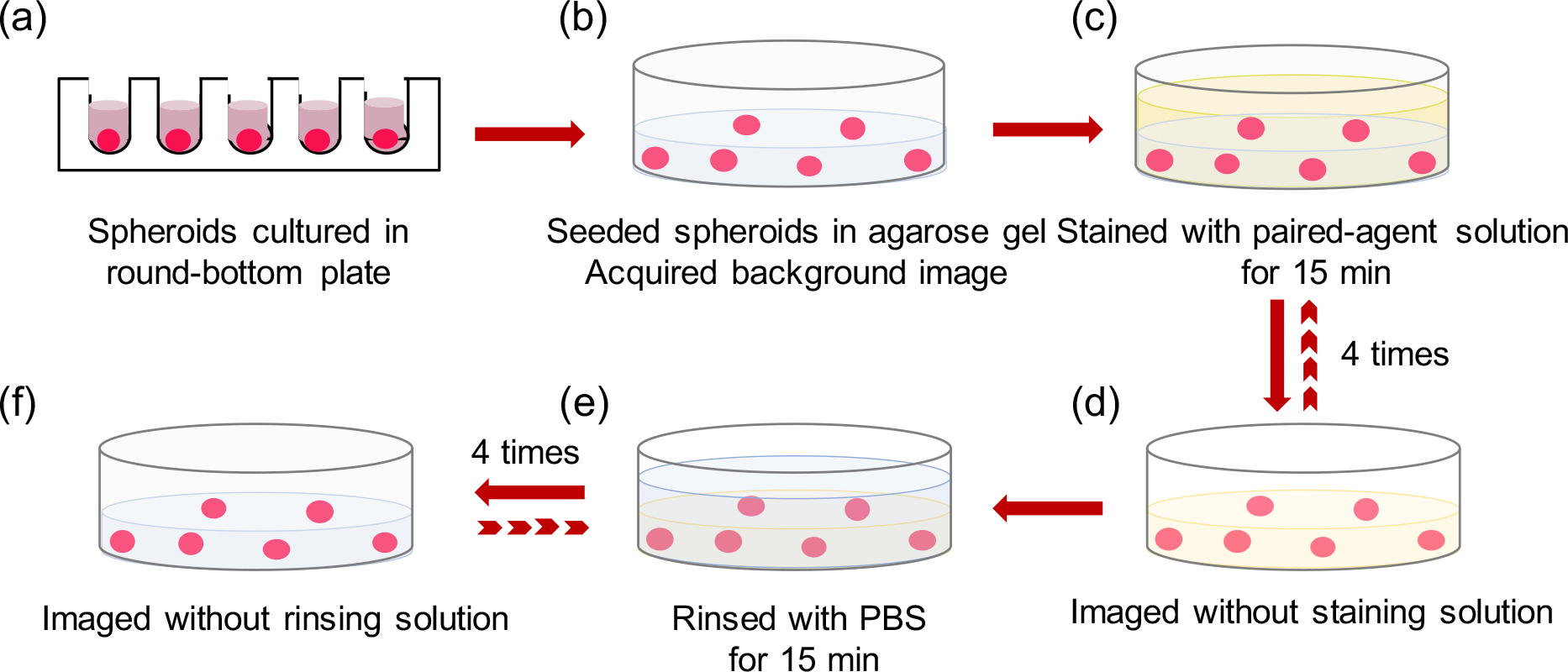
Stepwise illustration of 3-D tumor spheroids staining and rinsing protocol in a well plate. (a)Tumor spheroids were formed in the round-bottom plate, (b) then embedded in 0.3% agarose gel and a background image was acquired to account for autofluorescence. (c) 5-mL mixed imaging agent solution was added to stain encapsulated spheroids for 1 h. (d) During the staining process, staining solution supernatant was removed and the fluorescence of the targeted agent and control agent were recorded at 15-min intervals. (e) Spheroids were then followed by 1-h rinsing with 5-mL PBS; rinsing solution was removed every 15 min and the fluorescence images were acquired (f).
2.4. Image post-processing
Images were analyzed using in-house code written in MATLAB (R2018b, Mathwork, Natick, MA) on a pixel-by-pixel basis. Pre-injection images were subtracted from all post-injection images (following motion correction using rigid body translation/rotation in built in MATLAB functions) to remove the effects of background autofluorescence. A circular region-of-interest (ROI) of ~600 μm diameter was selected over tumor spheroids, visible on the white-light Pearl images. Spheroids embedded within 5 mm off the edge of the wells were excluded from analyses owing to observed distortion of the fluorescence by the well wall. Fluorescence signal of all analyzed tumor spheroids in the same well were averaged at each time point and the mean signals were used to represent entire wells.
2.5. Estimating kinetic parameters
The kinetic parameters of and , the rates of exchange of the imaging agent between , the staining solution concentration, to , the interstitial free space, were extrapolated by fitting the analytical solution of a one-compartment model (Eq. 4) to the temporal fluorescence signal of the control agent in each well, independently:
| (4) |
where represents the uptake/retention of the control agent in the region of interest; and is the correction factor relating detected control agent signal to control agent concentration, which was estimated by measuring the fluorescence signal of the stock solution.
By assuming similarity between and —the transport kinetics between the staining solution and free space of the control agent and targeted agent to be the same (i.e., )—the solutions to the differential equations in Eq. 2 can be written as follows:
| (5) |
where represents the measured fluorescence signal curve of the targeted agent in the region of interest, and represents the correction factor relating detected targeted agent signal to targeted agent concentration.
The rate of association and dissociation of the targeted agent, and respectively, were determined by nonlinear least squares fitting of Eq. 5 to measured uptake/retention of targeted agent in each well. The resulting kinetic parameters were then used in mathematical model simulations using Eq. 1 & 2 and compared to the experimental data.
2.6. Statistical analyses
A repeated-measures, two-way ANOVA, with cancer-type as a between-subjects factor and time as a within-subjects factor, was used to identify statistically significant effects of temporal uptake and washout kinetics of the targeted agent amongst different EGFR expression cancer types. To determine statistical significance between groups, one-way ANOVA was performed, followed by Tukey’s post-test for pairwise and multiple groups comparison. Linear regression was employed to evaluate the strength of the correlation between experimental data and model-simulated data. Statistical significance was based on . All data are presented as mean ± sd.
3. Results
3.1. In vitro 3-D spheroids imaging
The representative fluorescence images of the control and targeted imaging agents (including the pre-staining images and subsequent images during the staining and rinsing process) in a well with high levels of EGFR expression cell spheroids are presented in Fig. 3(a). Images acquired in the 700-nm channel in the Pearl system are shown in red and the fluorescence in the 800-nm channel in the Pearl system are shown in green. A background subtraction algorithm was applied to both imaging agent uptake curves to correct for background and autofluorescence signals. Fluorescence signal of individual spheroids were averaged and the mean fluorescence from both IRDye-700DX carboxylate and ABY-029 were used to analyze their kinetics over time for each well. Fig 3(b) demonstrates the temporal average uptake and washout kinetics of ABY-029 in the spheroid-embedded wells ( wells for each group) for all cancer groups with different EGFR expression levels (High: A431, Moderate: MDA-MB-231, Low: BT-474, negative control: SW620), as well as the uptake of the control imaging agent. The results display the varied enhanced retention of the targeted agent in different cancer cell lines, where a proportionally greater separation was observed with the cancer type that expressed more EGFR between all cancer types. A repeated-measures ANOVA with cancer-type as a between-subjects factor and time as a within-subjects factor demonstrated that the temporal uptake of ABY-029 was significantly different among all cancer types ().
Figure 3.
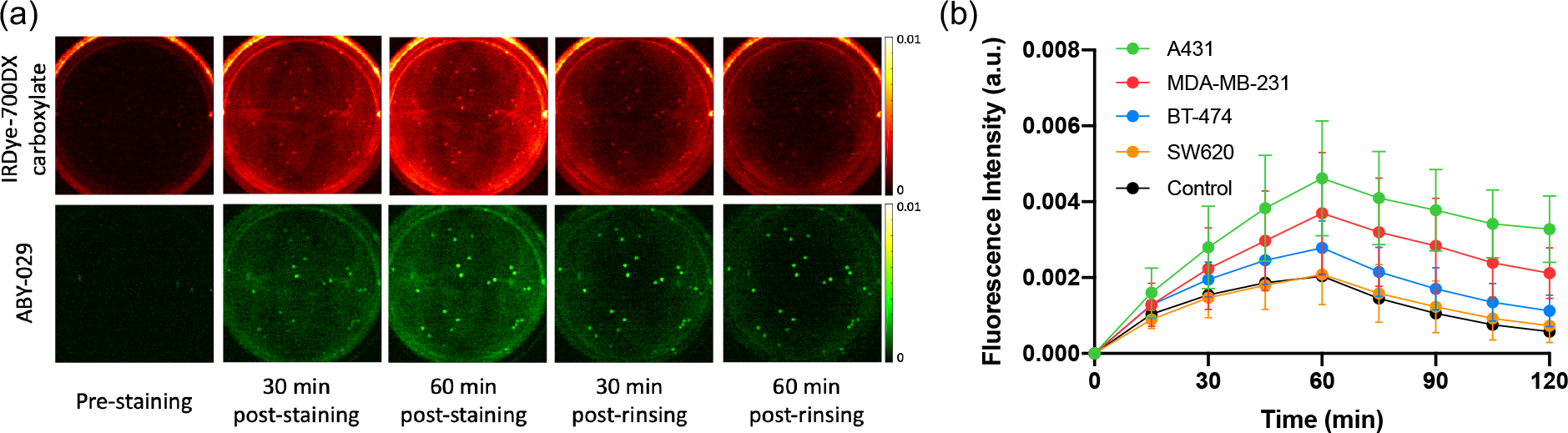
Kinetics of ABY-029 and IRDye-700DX carboxylate over time during staining and rinsing process. (a) Fluorescence images of a representative well of high EGFR expression cancer cell spheroids in the staining and rinsing procedure. Pre-staining images were acquired to evaluate background signal and autofluorescence. The top row represents the temporal signal acquired at 700 nm channel (shown in red) indicating the IRDye-700DX carboxylate fluorescence, and the bottom row represents the temporal signal acquired at 800 nm channel (shown in green) indicating ABY-029 fluorescence. (b) The average (mean ± sd) fluorescence for both imaging agents in n=5 wells for each cancer type as a function of time.
3.2. Kinetic parameters estimation
Kinetic parameters estimated by nonlinear least square fitting of Eq. (4) and (5) to the in vitro data are shown in Figure 4. Fig 4(a) presents the estimated values as the mean± sd: and , while Fig 4(b) shows the estimated values were and for A431 (high), MDA-MB-231 (moderate), BT-474 (low) and SW620 (control), respectively. No significant differences were observed among the different cancer groups with these two parameters, which govern the transit of imaging agent between staining/rinsing solution and interstitial free space. Fig 4(c) presents the estimated association constant , with and with decreasing EGFR expression levels. The average from the high EGFR expression group (A431) was significantly higher than moderate (MDA-MB-231) and low (BT-474) EGFR expression groups and the control group (SW620), while the value from the moderate group was significantly higher than the control group . No statistical differences were observed among different groups in disassociation constant estimation, with of and for groups of A431, MDA-MB-231, BT-474 and SW620, respectively.
Figure 4.
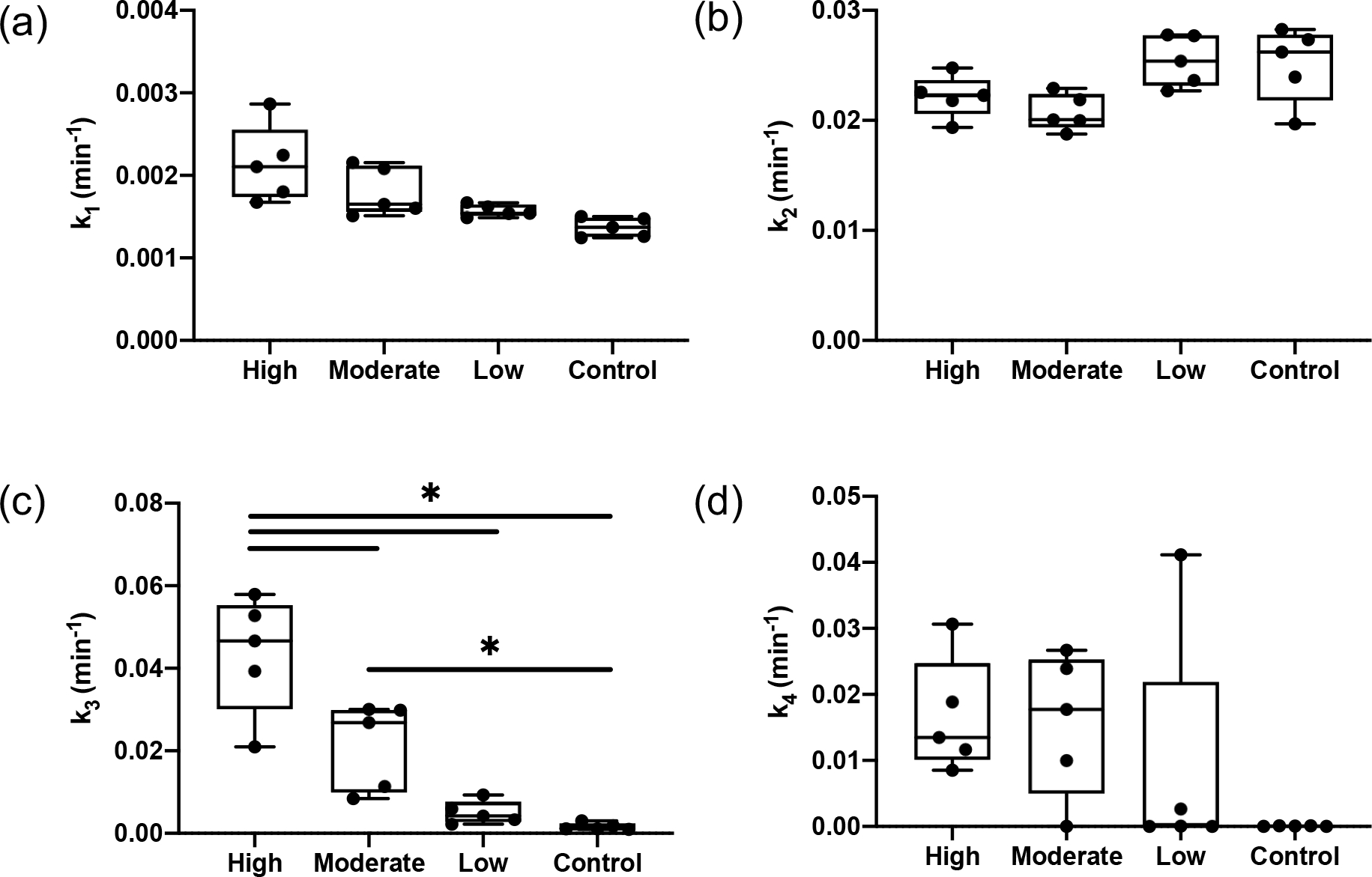
Estimated kinetic parameters for different EGFR expression cancer-type groups (High: A431, Moderate:MDA-MB-231, Low: BT-474, Control: SW620). (a) - rate constant governing the imaging agent transfer from staining/rinsing solution to interstitial space. (b) - rate constant governing the imaging agent transfer from interstitial space out to staining/rinsing solution. (c) - rate constant governing the targeted agent from interstitial space associate with its specific receptor. (d) - rate constant governing the separation of the bound imaging agent back into the interstitial space.
Figure 5 presents the validation of the model-estimated binding kinetic parameters. The estimated association rate constants, , were compared with the epidermal growth factor receptors per cell for all different cancer types. The human epidermoid carcinoma A431, which expresses about 2 × 106 receptors/cells, represented the high level of EGFR expression cancer group (30), with descending order of EGFR expression in MDA-MB-231 and BT-474, while SW620 represented the no measurable receptor expression or control group (30; 31; 36). A statistically significant correlation () was observed between the association rate constant and the EGFR expression levels among the cell lines (Fig 5(a)). These results demonstrated the feasibility of the model estimation as the rate constant is theoretically proportional to the receptor concentration (28). The equilibrium disassociation constant was determined by the ratio of dissociation constant, , to association constant, , with consideration of receptor concentration for different cell lines with varied levels of EGFR expression (Fig 5(b)). The total concentration of receptors available, , was estimated by the spherical expression on the area of a 300-μm spheroid with the reported cell surface EGFR density (37). Fig 5(b) demonstrated no appreciable differences between the high EGFR expression group and the moderate EGFR expression group. Meanwhile, considerably smaller values of binding affinity, , were observed in the low receptor density group.
Figure 5.
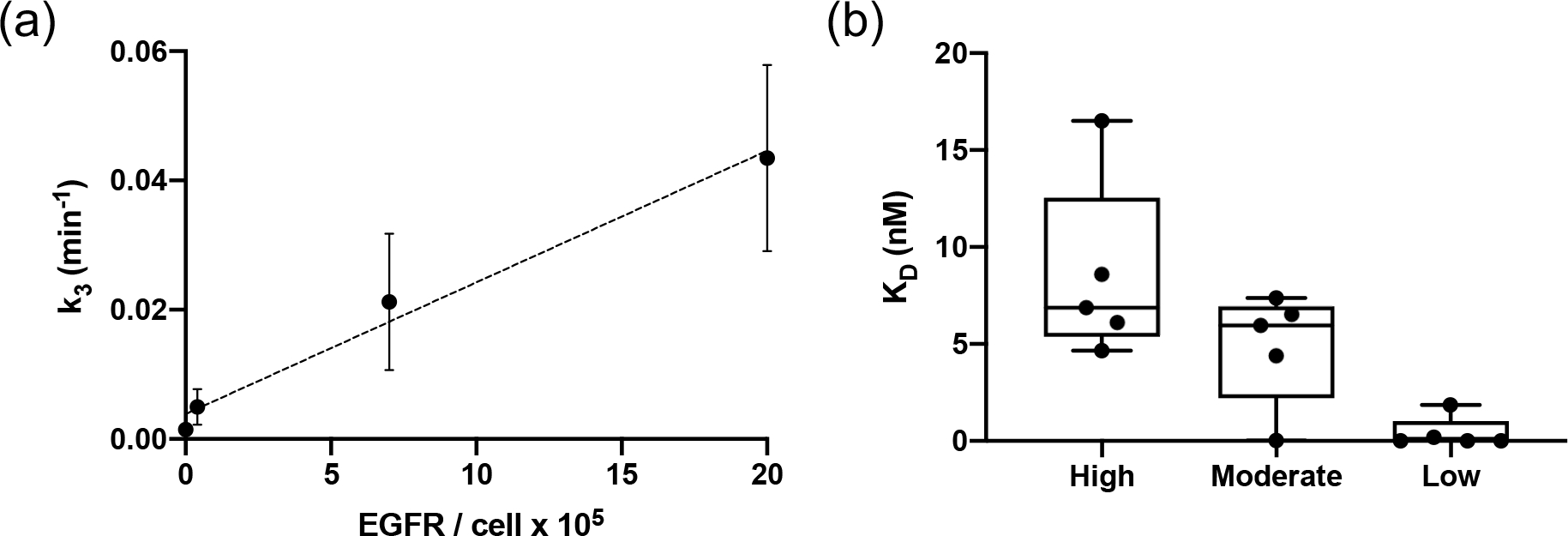
Validation of estimated kinetic parameters. (a) Presents the correlation of the average association rate constant of the A431, MDA-MB-231, BT-474 and SW620 groups to the corresponding EGFR expression level (). (b) Shows the comparison of estimated binding affinity with the model in high, moderate and low receptor expression groups.
3.3. Numerical simulations using estimated parameters
The model was further evaluated by simulating the concentration profile of the targeted and control agent using all estimated kinetic parameters from section 3.2 and compared with the in vitro experiments. Fig 6(a)–(d) shows the raw simulated data (shown in dashed lines) and experimental results (shown in solid lines) for both targeted agent (shown in green) and control agent (shown in red) with different cancer-type groups of A431, MDA-MB-231, BT-474 and SW620, respectively. The average experimental data was displayed with standard deviation ( wells for each cancer type), and the shaded regions indicate the standard deviation of the simulated data ( for each cancer type) based on the estimated kinetic parameters from the experimental data.
Figure 6.
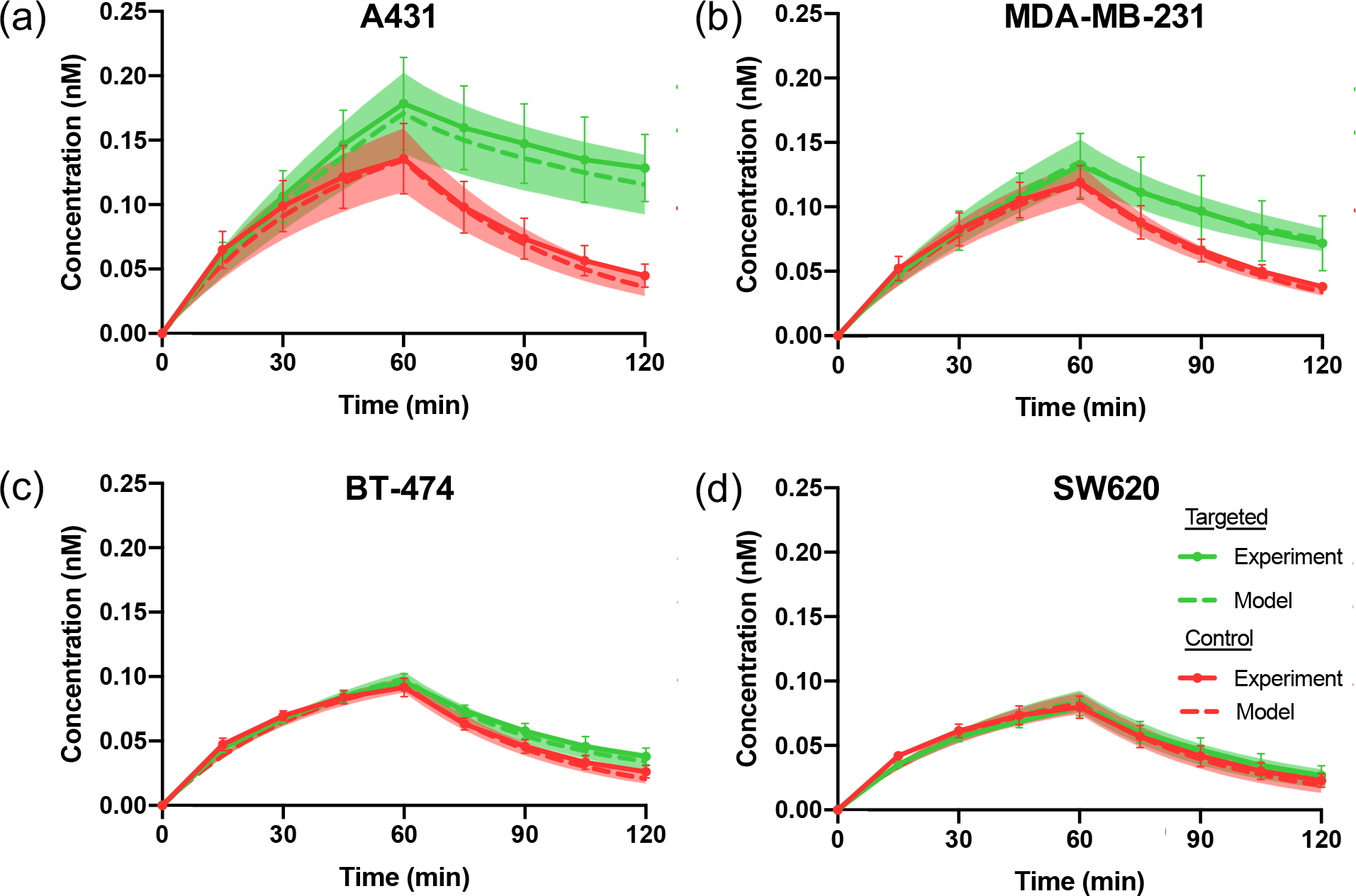
Comparison of model-simulated data and in vitro experimental data. Experimental concentrations of targeted agent (green) and control agent (red) are shown as solid lines, while model-simulated data are shown as dashed lines. Standard deviation is shown as error bars in the experimental data, and as the shaded region for the simulated data. (a) – (d) show the concentration profile comparison of experimental and simulated data in high EGFR expression cell line A431, moderate EGFR expression cell line MDA-MB-231, low expression of EGFR in BT-474 and negative control cell line SW620, respectively.
To validate the accuracy of estimated parameters, root-mean-square error and R-squared regression values were calculated to evaluate the correlation of the simulated data with the model and the experimental data presented in Table 1. The root-mean-square error remained small among different cancer-type groups with various EGFR expression levels, and the R-squared values were for all groups.
Table 1.
Correlations of in vitro experimental data and model-simulated data with fractional root-mean-square erro
| Root-mean-square error | R-squared | |
|---|---|---|
| A431 (High) | 0.009 ± 0.003 | 0.99 ±0.003 |
| MDA-MB-231 (Moderate) | 0.006 ±0.001 | 0.97 ±0.04 |
| BT-474 (Low) | 0.004 ± 0.0008 | 0.99 ±0.001 |
| SW620 (Control) | 0.003 ± 0.0009 | 0.99 ±0.001 |
4. Discussion
Evaluation of in vivo relevant binding kinetics and the rate constants of binding-site association and dissociation are critical for elucidating complexities of cancer diagnosis and treatment compared to more conventional measures of drug or imaging agent binding affinity on its own. At the molecular level, drug binding can be affected by the microenvironment surrounding the site of binding; therefore, the binding kinetics tend to provide more information to better evaluate the clinical efficacy, duration of action, therapeutic differentiation, and safety profiles of the drug (9). The significance of these two indicators has been recognized now and covered in different review articles (5; 9). Although notable efforts have been made, cancer drug development has a significantly higher attrition rate than other therapeutic areas (38). Most current techniques of evaluating drug binding in vitro are costly and limited to two-dimensional biological studies that fail to recapitulate the tumor complexity profiles. In this study, a feasible methodology of modeling and estimating binding kinetics with paired-agent molecular imaging approaches in 3-D tumor spheroid tissue culture is presented. The results demonstrated the methodology’s capability of quantifying the parameters with a sufficient translational tumor model.
In vitro 3-D tumor cell culture model was used to provide a more complex and realistic environment of three-dimensional extracellular platform, which is much closer to clinical expression profiles, to perform the binding assays. To fully present their physiological form and function, the 3-D cancer spheroids were embedded in agarose gel. A softer concentration of 0.3% agarose in saline was selected to facilitate the imaging agent diffusion and minimize the external forces on the tumor spheroids. However, this gel-encapsulated technique created a challenge that made the direct measurement of kinetic rate constants of the compound interaction impossible. The paired-agent methodology has been applied to overcome this limitation by employing a control imaging agent to account for the diffusion and nonspecific retention of the targeted agent (26). A one-compartment model was used to calculate the kinetic parameters and , which was used to estimate the exchange rate between the staining/rinsing solution and interstitial space-embedded gel. Fig 4(a) and (b) show that each estimated parameter exhibited low variance with no significant differences among different groups. The consistency of and across multiple cell lines make it reliable to further investigate the binding kinetics in the two-compartment model.
This model required that targeted and control agents share identical kinetics in the absence of binding. An anti-EGFR Affibody imaging agent, ABY-029, was selected to be the targeted agent and IRDye-700DX carboxylate was used as the control agent. The experiments were carried out in blank wells ( wells) with 0.3% agarose gel containing no cancer spheroids. and the kinetic curves showed a strong correlation of both imaging agents () over the entire staining and rinsing procedure (results not shown). The equivalence of rate constants and in control agent and targeted agent allowed the binding kinetics to be modelled with the two-compartment model. This is because the model relies on the assumptions that the different levels of agent-target binding to encapsulated spheroids have no measurable effects on the kinetic rate constants and (i.e. ), and that the ratio of bound imaging agent concentration to free imaging agent concentration remains relatively constant at all imaging times. These assumptions were tested by using the approximate kinetic parameters to the full kinetic model and comparing them with the experimental data (Table 1). This resulted in less than 1% in averaged root-mean-square error amongst all cancer types with different levels of EGFR expression.
Fig 4(c) shows the estimated association rate constant, , correlated well with the level of EGFR expression in the cell line tested, which is expected as is posited to be the product of the total available receptor concentration () on the on-rate of binding, (28). Plotting the association rate constant, , against the epidermal growth factor receptors per cell for all different cancer types (Fig 5 (a)), a strong correlation was observed (), which further elucidated that the changes in were proportional to the receptor concentration. Compared to high and moderate EGFR expression groups, the average estimated dissociation rate constant, , displayed smaller values in the low expression group with considerable variation, and over two-orders of magnitude smaller values were observed in the control group presented in Fig 4(d). The discrepancy in the estimations of dissociation rate constant, , may be owing to the low association rate constant, , as the model works by nonlinear least squares fitting of the measured signal of the imaging agent. Although the paired-agent approach possesses a highly sensitive profile that was able to detect enhanced retention in the cell lines known for low EGFR expression, the low values of estimation in were compensated by low values of estimation in . These results suggested that receptor-overexpressing cell lines are preferred for optimal use of this binding kinetic modeling.
Alternatively, the variability in could be linked to variability in levels of imaging agent cell internalization amongst the tumor cell lines. Some work has demonstrated that the affibodies used in this student can exhibit up to 20% cell internalization over 1 h of incubation in A431 cells (39). This is likely an overestimate of the likelihood for internalization since A431 is a very high over-expressor of EGFR. Moreover, this study was carried out in 2D cell culture where cell expansion over the dish leads to high surface-area-to-volume, likely amplifying internalization potential over more clinically relevant 3-D culture methods as used here. According to simulations, a 20% internalization over 1 h of binding, with no leakage back out of the cell during the 1 h rinse stage in this work, would have led to a predicted underestimation of 20% on the parameter measured, with no effect appreciable effect on the parameter. Assuming internalization should scale with receptor concentration, the observation that , was similar in high and moderate expression groups (A431 and MDA-MB-231, respectively), suggests that internalization may not have been a dominating factor in these studies. Estimated (analogous to in vitro ) for ABY-029 to EGFR were lower than the values obtained in surface plasmon resonance studies, which were reported as 0.11 min−1 (35), compared to the ~0.02 min−1 reported here, which could point to significant internalization. However, several studies have suggested that the in vivo dissociation rate, , is expected to be smaller than in vitro (28). Further investigation is necessary to fully determine the effect internalization will have on this methodology. One option, for instance, to minimize effects of internalization, could be to reduce staining and rinsing times, considerably. This work represents a preliminary demonstration of the methodology and timing of staining and rinsing was not optimized.
was determined by the ratio of to with consideration of for different cell types. was estimated by the spherical expression on the surface area of a 300-μm spheroid with the reported cell surface EGFR density. For instance, the averaged A431 cell surface EGFR density reported as , was applied to calculate the total receptor concentrations on the surface area of a 300-μm diameter spheroid. The averaged determination of ABY-029 to EGFR in A431 and MDA-MB-231 was found to be , where a study of anti-EGFR Affibody molecules was found to have values in the 5–10nM range (40). However, further simulation studies are necessary to validate this simplified estimation and accurately determine the total receptor expression in tumor spheroids.
Despite concerns about underestimation of , it is interesting that our in vitro estimates of (equilibrium disassociation constant) match well with in vivo methods, because of proportionally decreased estimates of . The smaller values indicated that the estimated parameters with our model have potential to represent the in vivo profiles of biological activity and functions more accurately. However, a more comprehensive model with implementation of characterized cellular internalization is of interest for future work.
Binding site barrier effects, where high rates of binding at the spheroid surface can impede apparent diffusion of the targeted agent into the spheroid (41; 42), could also affect the results of the methodology to correlate to 2-D in vitro methods; however, initial diffusion modeling simulations suggest very little influence of binding-site barrier on ABY-029 diffusion in small tumor spheroids (43). However, in cases where binding-site barrier were to influence ABY-029 diffusion, this could be seen as a further restriction of the number of “available” drug or imaging agent binding sites that would be realistic for in vivo cases but not captured by in vitro methods. Simulations that aim to guide optimization of drug dosing and scheduling or imaging agent dosing and time-of-imaging would benefit from methods like that presented in this paper that provide a more equivalent depiction of in vivo apparent binding kinetics.
In this study, we presented a low-cost, novel, and promising in vitro methodology to model binding kinetics on 3-D cancer spheroids using paired-agent imaging approaches. There are several significant preclinical implications that could benefit from this work. For cancer diagnosis, the estimated binding kinetics can provide in vivo kinetic uptake curves of targeted agent by incorporation of plasma input curves of the imaging agent to evaluate the optimal concentration and imaging time point to achieve the highest contrast-to-noise ratio (CNR) in tumor discrimination (44). Moreover, the kinetic parameters could guide drug development, better evaluating the drug-to-target-binding effects in the temporal physiological in vivo environment, and therefore optimizing drug efficiency. To better simulate and characterize the biological information of human tumor, multicellular spheroid systems can be applied to this model to resemble cell heterogeneity, tumor vasculature and other physiological conditions operative in 3D microenvironments (45–47).
5. Conclusion
This study demonstrated the potential of a novel binding kinetic model paired with a 3-D spheroid tumor cell culture platform to estimate two favorable kinetic parameters: association and dissociation rate constants. This cost-effective and robust approach will allow the development of cancer diagnosis and therapeutics to identify and optimize the efficacy of the targeted molecules in a more clinically relevant model.
Funding
This work has been funded from National Science Foundation (1653627), Nayar Prize at Illinois Tech and National Cancer Institute NCI R01 CA167413 for the synthesis of ABY-029.
Abbreviations, symbols, and terminology:
- ABY-029
A fluorescently labeled antibody mimetic that is targeted to epidermal growth factor receptor
- BLI
Biolayer interferometry
- ,
Bound concentration of the control agent
- ,
Free concentration of the control agent
- ,
Concentration of the control agent in the staining solution
- ,
Bound concentration of the targeted agent
- ,
Free concentration of the targeted agent
- ,
Concentration of the targeted agent in the staining solution
- EGFR
Epidermal growth factor receptor
- GLP
Good laboratory practices
- IRDye 700DX
Commercial fluorescent molecular with peak emission near 700 nm (LI-COR Biosciences, Lincoln, NE)
- IRDye 800CW
Commercial fluorescent molecular with peak emission near 800 nm (LI-COR Biosciences, Lincoln, NE)
- ,
Association constant, “affinity”
- ,
Dissociation constant
- ,
Rate constant governing transport of the control agent from staining solution to tissue culture
- ,
Rate constant governing transport of the control agent from tissue culture to staining solution
- ,
Rate constant governing transport of the targeted agent from staining solution to tissue culture
- ,
Rate constant governing transport of the targeted agent from tissue culture to staining solution
- ,
Rate constant governing in vivo receptor binding
- ,
Rate constant governing
- SPR
Surface plasmon resonance
Footnotes
Conflict of interest
Dr. Samkoe reports grants from National Cancer Institute, during the conduct of the study; and ABY-029 was manufactured under National Cancer Institute NCI R01 CA167413 grant as an academic-industrial partnership between Dartmouth College, Affibody AB, and LI-COR Biosciences, Inc.
Citation diversity statement
Recent work in several fields of science has identified a bias in citation practices such that papers from women and other minority scholars are undercited relative to the number of papers in the field. We recognize this bias and have worked diligently to ensure that we are referencing appropriate papers with fair gender and racial author inclusion.
References
- 1.Hughes JP, Rees S, Kalindjian SB, Philpott KL. 2011. Principles of early drug discovery. British journal of pharmacology 162:1239–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth CW, Gibbs SL. 2020. Fluorescence Image-Guided Surgery - a Perspective on Contrast Agent Development. Proc SPIE Int Soc Opt Eng 11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hernot S, van Manen L, Debie P, Mieog JSD, Vahrmeijer AL. 2019. Latest developments in molecular tracers for fluorescence image-guided cancer surgery. Lancet Oncol 20:e354–e67 [DOI] [PubMed] [Google Scholar]
- 4.Ma W, Yang L, He L. 2018. Overview of the detection methods for equilibrium dissociation constant KD of drug-receptor interaction. Journal of pharmaceutical analysis 8:147–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Copeland RA, Pompliano DL, Meek TD. 2006. Drug–target residence time and its implications for lead optimization. Nature reviews Drug discovery 5:730–9 [DOI] [PubMed] [Google Scholar]
- 6.Pan AC, Borhani DW, Dror RO, Shaw DE. 2013. Molecular determinants of drug–receptor binding kinetics. Drug discovery today 18:667–73 [DOI] [PubMed] [Google Scholar]
- 7.Tiwary P, Limongelli V, Salvalaglio M, Parrinello M. 2015. Kinetics of protein–ligand unbinding: Predicting pathways, rates, and rate-limiting steps. Proceedings of the National Academy of Sciences 112:E386–E91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Núñez S, Venhorst J, Kruse CG. 2012. Target–drug interactions: first principles and their application to drug discovery. Drug discovery today 17:10–22 [DOI] [PubMed] [Google Scholar]
- 9.Swinney DC. 2009. The role of binding kinetics in therapeutically useful drug action. Current opinion in drug discovery & development 12:31. [PubMed] [Google Scholar]
- 10.Scarano S, Scuffi C, Mascini M, Minunni M. 2010. Surface plasmon resonance imaging (SPRi)-based sensing: a new approach in signal sampling and management. Biosens Bioelectron 26:1380–5 [DOI] [PubMed] [Google Scholar]
- 11.Huber W, Mueller F. 2006. Biomolecular interaction analysis in drug discovery using surface plasmon resonance technology. Current pharmaceutical design 12:3999–4021 [DOI] [PubMed] [Google Scholar]
- 12.Nordin H, Jungnelius M, Karlsson R, Karlsson OP. 2005. Kinetic studies of small molecule interactions with protein kinases using biosensor technology. Analytical biochemistry 340:359–68 [DOI] [PubMed] [Google Scholar]
- 13.Concepcion J, Witte K, Wartchow C, Choo S, Yao D, et al. 2009. Label-free detection of biomolecular interactions using BioLayer interferometry for kinetic characterization. Combinatorial chemistry & high throughput screening 12:791–800 [DOI] [PubMed] [Google Scholar]
- 14.Shah NB, Duncan TM. 2014. Bio-layer interferometry for measuring kinetics of protein-protein interactions and allosteric ligand effects. JoVE (Journal of Visualized Experiments):e51383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karlsson R, Fält A. 1997. Experimental design for kinetic analysis of protein-protein interactions with surface plasmon resonance biosensors. Journal of immunological methods 200:121–33 [DOI] [PubMed] [Google Scholar]
- 16.Fennema E, Rivron N, Rouwkema J, van Blitterswijk C, de Boer J. 2013. Spheroid culture as a tool for creating 3D complex tissues. Trends Biotechnol 31:108–15 [DOI] [PubMed] [Google Scholar]
- 17.Zanoni M, Piccinini F, Arienti C, Zamagni A, Santi S, et al. 2016. 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Sci Rep 6:19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sant S, Johnston PA. 2017. The production of 3D tumor spheroids for cancer drug discovery. Drug Discov Today Technol 23:27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He YJ, Young DA, Mededovic M, Li K, Li C, et al. 2018. Protease-Sensitive Hydrogel Biomaterials with Tunable Modulus and Adhesion Ligand Gradients for 3D Vascular Sprouting. Biomacromolecules 19:4168–81 [DOI] [PubMed] [Google Scholar]
- 20.He YJ, Santana MF, Moucka M, Quirk J, Shuaibi A, et al. 2020. Immobilized RGD concentration and proteolytic degradation synergistically enhance vascular sprouting within hydrogel scaffolds of varying modulus. J Biomater Sci Polym Ed 31:324–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Torres VC, Tichauer KM. 2018. Noninvasive detection of cancer spread to lymph nodes: A review of molecular imaging principles and protocols. J Surg Oncol 118:301–14 [DOI] [PubMed] [Google Scholar]
- 22.Li C, Torres VC, He Y, Xu X, Basheer Y, et al. 2021. Intraoperative Detection of Micrometastases in Whole Excised Lymph Nodes Using Fluorescent Paired-Agent Imaging Principles: Identification of a Suitable Staining and Rinsing Protocol. Mol Imaging Biol 23:537–49 [DOI] [PubMed] [Google Scholar]
- 23.Torres VC, Li C, He Y, Sinha L, Papavasiliou G, et al. 2019. Angular restriction fluorescence optical projection tomography to localize micrometastases in lymph nodes. J Biomed Opt 24:1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torres VC, Li C, Zhou W, Brankov JG, Tichauer KM. 2021. Characterization of an angular domain fluorescence optical projection tomography system for mesoscopic lymph node imaging. Appl Opt 60:135–46 [DOI] [PubMed] [Google Scholar]
- 25.Torres VC, Li C, Brankov JG, Tichauer KM. 2021. Model-based system matrix for iterative reconstruction in sub-diffuse angular-domain fluorescence optical projection tomography. Biomed Opt Express 12:1248–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tichauer KM, Samkoe KS, Sexton KJ, Hextrum SK, Yang HH, et al. 2012. In vivo quantification of tumor receptor binding potential with dual-reporter molecular imaging. Mol Imaging Biol 14:584–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lammertsma A, Bench C, Hume S, Osman S, Gunn K, et al. 1996. Comparison of methods for analysis of clinical [11C] raclopride studies. Journal of Cerebral Blood Flow & Metabolism 16:42–52 [DOI] [PubMed] [Google Scholar]
- 28.Innis RB, Cunningham VJ, Delforge J, Fujita M, Gjedde A, et al. 2007. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab 27:1533–9 [DOI] [PubMed] [Google Scholar]
- 29.Wikstrand CJ, McLendon RE, Friedman AH, Bigner DD. 1997. Cell surface localization and density of the tumor-associated variant of the epidermal growth factor receptor, EGFRvIII. Cancer research 57:4130–40 [PubMed] [Google Scholar]
- 30.Anido J, Matar P, Albanell J, Guzmán M, Rojo F, et al. 2003. ZD1839, a specific epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor, induces the formation of inactive EGFR/HER2 and EGFR/HER3 heterodimers and prevents heregulin signaling in HER2-overexpressing breast cancer cells. Clinical Cancer Research 9:1274–83 [PubMed] [Google Scholar]
- 31.Petrulli JR, Sullivan JM, Zheng M-Q, Bennett DC, Charest J, et al. 2013. Quantitative analysis of [11C]-erlotinib PET demonstrates specific binding for activating mutations of the EGFR kinase domain. Neoplasia 15:1347–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li C, Xu X, Basheer Y, He Y, Sattar HA, et al. Paired-agent fluorescent imaging to detect micrometastases in breast sentinel lymph node biopsy: experiment design and protocol development. Proc. Advanced Biomedical and Clinical Diagnostic and Surgical Guidance Systems XVI, 2018, 10484:1048402: International Society for Optics and Photonics [Google Scholar]
- 33.Henderson ER, Werth P, Xu X, Jarvis LA, Paulsen KD, et al. 2022. Epidermal growth factor-targeted fluorescence is unaffected by standard neoadjuvant therapies in human sarcomas. Proc SPIE Int Soc Opt Eng 119430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samkoe KS, Hull S, Elliott J, Sardar HS, Gunn J, et al. 2020. Perspectives on the Phase 0 clinical trial of microdose administration of ABY-029 for fluorescence guided surgery: Stability testing. Proc SPIE Int Soc Opt Eng 11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samkoe KS, Gunn JR, Marra K, Hull SM, Moodie KL, et al. 2017. Toxicity and Pharmacokinetic Profile for Single-Dose Injection of ABY-029: a Fluorescent Anti-EGFR Synthetic Affibody Molecule for Human Use. Mol Imaging Biol 19:512–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fitzpatrick SL, LaChance MP, Schultz GS. 1984. Characterization of epidermal growth factor receptor and action on human breast cancer cells in culture. Cancer research 44:3442–7 [PubMed] [Google Scholar]
- 37.Zhang F, Wang S, Yin L, Yang Y, Guan Y, et al. 2015. Quantification of epidermal growth factor receptor expression level and binding kinetics on cell surfaces by surface plasmon resonance imaging. Analytical chemistry 87:9960–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ocana A, Pandiella A, Siu LL, Tannock IF. 2011. Preclinical development of molecular-targeted agents for cancer. Nature reviews Clinical oncology 8:200–9 [DOI] [PubMed] [Google Scholar]
- 39.Göstring L, Chew MT, Orlova A, Höidén-Guthenberg I, Wennborg A, et al. 2010. Quantification of internalization of EGFR-binding Affibody molecules: Methodological aspects. Int J Oncol 36:757–63 [DOI] [PubMed] [Google Scholar]
- 40.Friedman M, Orlova A, Johansson E, Eriksson TL, Höidén-Guthenberg I, et al. 2008. Directed evolution to low nanomolar affinity of a tumor-targeting epidermal growth factor receptor-binding affibody molecule. Journal of molecular biology 376:1388–402 [DOI] [PubMed] [Google Scholar]
- 41.Singh AP, Guo L, Verma A, Wong GG, Thurber GM, Shah DK. 2020. Antibody Coadministration as a Strategy to Overcome Binding-Site Barrier for ADCs: a Quantitative Investigation. AAPS J 22:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thurber GM, Schmidt MM, Wittrup KD. 2008. Factors determining antibody distribution in tumors. Trends Pharmacol Sci 29:57–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kayaalp Nalbant E, Rounds C, Sadeghipour N, Meng B, Folaron MR, et al. 2020. A paired-agent fluorescent molecular imaging strategy for quantifying antibody drug target engagement in. Proc SPIE Int Soc Opt Eng 11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadeghipour N, Rangnekar A, Folaron MR, Strawbridge RR, Samkoe KS, et al. 2020. Prediction of optimal contrast times post-imaging-agent-administration to inform personalized fluorescence-guided surgery. J Biomed Opt Accepted [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin RZ, Chang HY. 2008. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnology Journal: Healthcare Nutrition Technology 3:1172–84 [DOI] [PubMed] [Google Scholar]
- 46.Kunz-Schughart LA, Freyer JP, Hofstaedter F, Ebner R. 2004. The use of 3-D cultures for high-throughput screening: the multicellular spheroid model. Journal of biomolecular screening 9:273–85 [DOI] [PubMed] [Google Scholar]
- 47.Kunz-Schugart LA, Kreutz M, Knuechel R. 1998. Multicellular spheroids: a three-dimensional in vitro culture system to study tumour biology. International journal of experimental pathology 79:1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]


