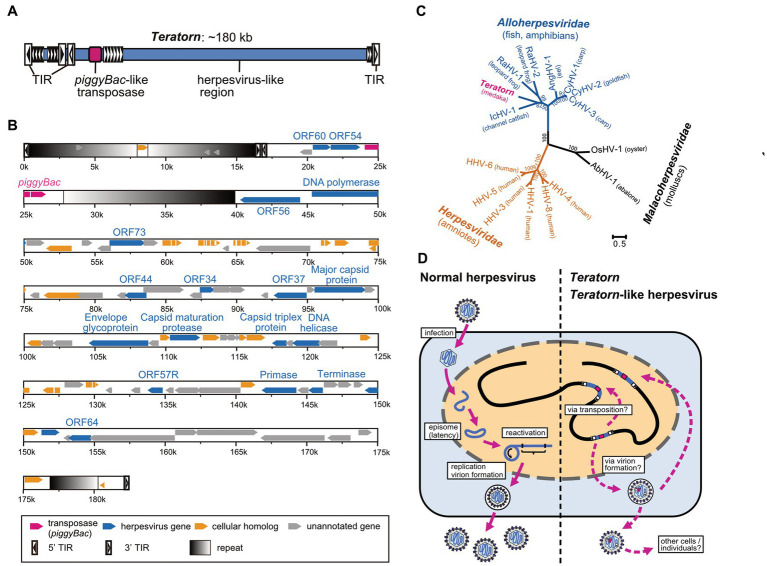
Figure 2.
Sequence characteristics of Teratorn and its proposed life cycle. (A) The overall structure of Teratorn. (B) Gene map of the subtype 1 Teratorn copy. Predicted genes are classified into four categories depicted by colored arrowheads; magenta, PiggyBac-like transposase gene; blue, herpesvirus-like genes; yellow, cellular homologs; gray, unannotated genes. Terminal inverted repeats (TIRs) of PiggyBac-like transposon are depicted by squares with black-to-white gradients. (C) Phylogeny of Teratorn and representative herpesvirus species, based on the maximum-likelihood analysis. Amino acid sequences of the DNA packaging terminase, the only gene confidently conserved among all herpesvirus species, were used. Bootstrap values of branching are indicated at the nodes. (D) Comparison of life cycles of normal herpesviruses (left) and Teratorn (right).