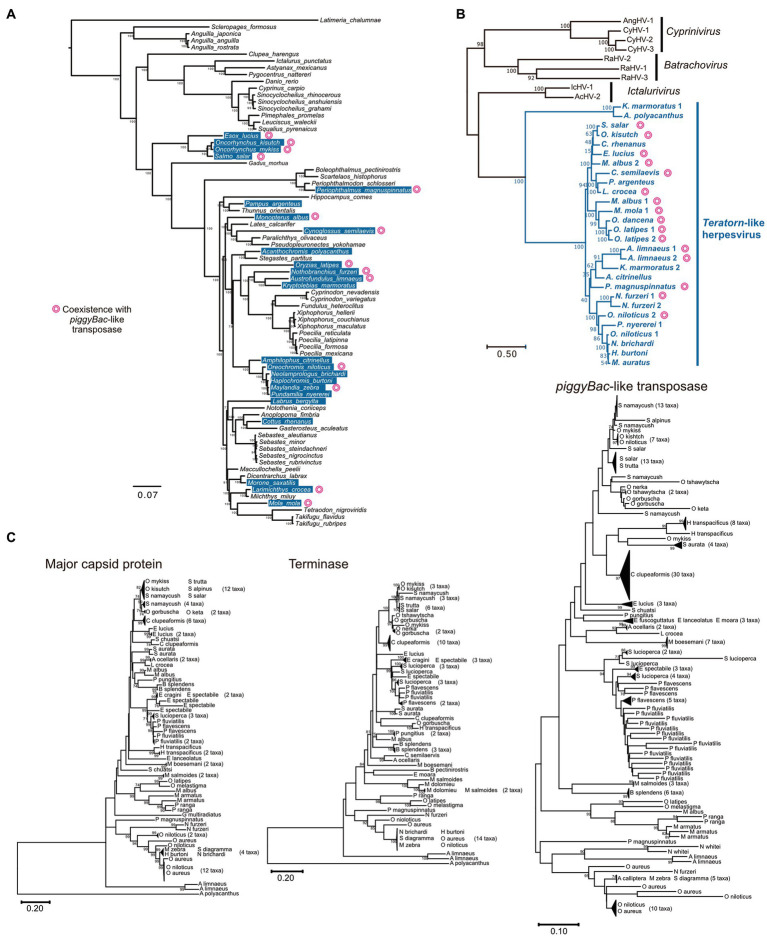
Figure 3.
Widespread distribution of Teratorn-like herpesviruses in teleosts. (A) Distribution of Teratorn-like herpesviruses in the genomes of teleost fish species. Species that seem to contain Teratorn-like herpesvirus are highlighted in blue (greater than nine of the 13 herpesvirus core genes, tblastn E-value <10−3). The phylogenetic tree was constructed by bayesian inference, based on the concatenated nucleotide sequence of nine nuclear genes (64). Species in which Teratorn-like herpesviruses are adjacent to piggyBac-like transposase genes are marked. (B) Phylogeny of Teratorn-like herpesviruses and other alloherpesvirus species, based on the maximum-likelihood analysis. The concatenated amino acid sequences of five herpesvirus genes (major capsid protein, DNA helicase, DNA polymerase, capsid triplex protein, and DNA packaging terminase) were used. (C) Maximum-likelihood trees based on the nucleotide sequences of major capsid protein, DNA packaging terminase, and piggyBac-like transposase genes inside re-screened Teratorn-like herpesvirus sequences. General time reversible model was used as a substitution model, considering evolutionary rate differences among sites by discrete Gamma distribution with five categories. Total of 2,878 positions (major capsid protein), 756 positions (DNA packaging terminase), and 558 positions (piggyBac-like transposase) were used in the final dataset. Nodes of phylogenetic trees were collapsed if the sequence similarity was greater than 98% for major capsid protein and terminase, and greater than 96% for piggyBac-like transposase. Non-collapsed trees are shown in Supplementary Figure 1. The scale bars represent the number of substitutions per site.