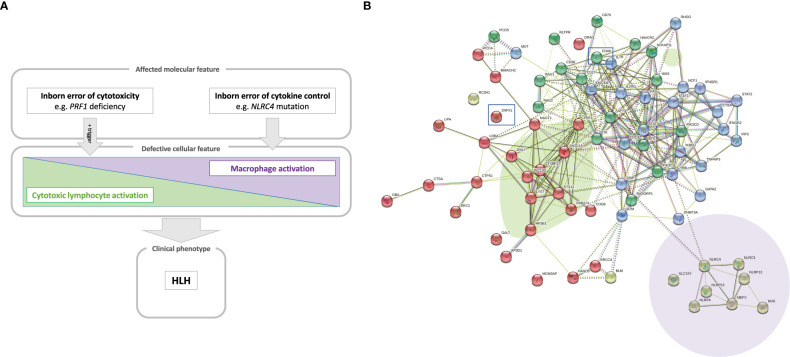
Figure 1.
(A) A model of the hyperinflammatory spectrum, ranging from errors of cytotoxicity to lack of cytokine control with macrophage activation. From the molecular point of view, HLH-associated gene mutations can be divided into inborn errors of cytotoxicity and inborn errors of cytokine control (upper panel). These genetic defects have consequences on the cellular level on a spectrum ranging from impaired lymphocyte cytotoxicity to macrophage activation. A trigger (e.g. a viral infection) for HLH is commonly found in the case of impaired lymphocyte cytotoxicity (middle panel). Both entities, inborn errors of cytotoxicity and inborn errors of cytokine control, may converge clinically to HLH (lower panel). (B) Genetic determinants of HLH and hyperinflammatory diseases. HLH-associated genes identified by searching the literature for reported cases are shown with their known and predicted protein-protein interaction network linked graphically using the STRING database and algorithm (182). The green oval highlights genes whose products are involved in granule-mediated cytotoxicity. Genes whose products interact with NLRC4 are highlighted by the mauve oval. HLH has also been described to occur in chronic granulomatous disease (genes marked in dark blue in the upper right corner) and (severe) combined immunodeficiency (genes in the upper left corner, such as IL2RG, ZAP70 and IL7RA). Two newly reported molecular causes, CD48 and ZNFX1, are highlighted (by squares), which we position here within the pathogenic pathways that lead to HLH.