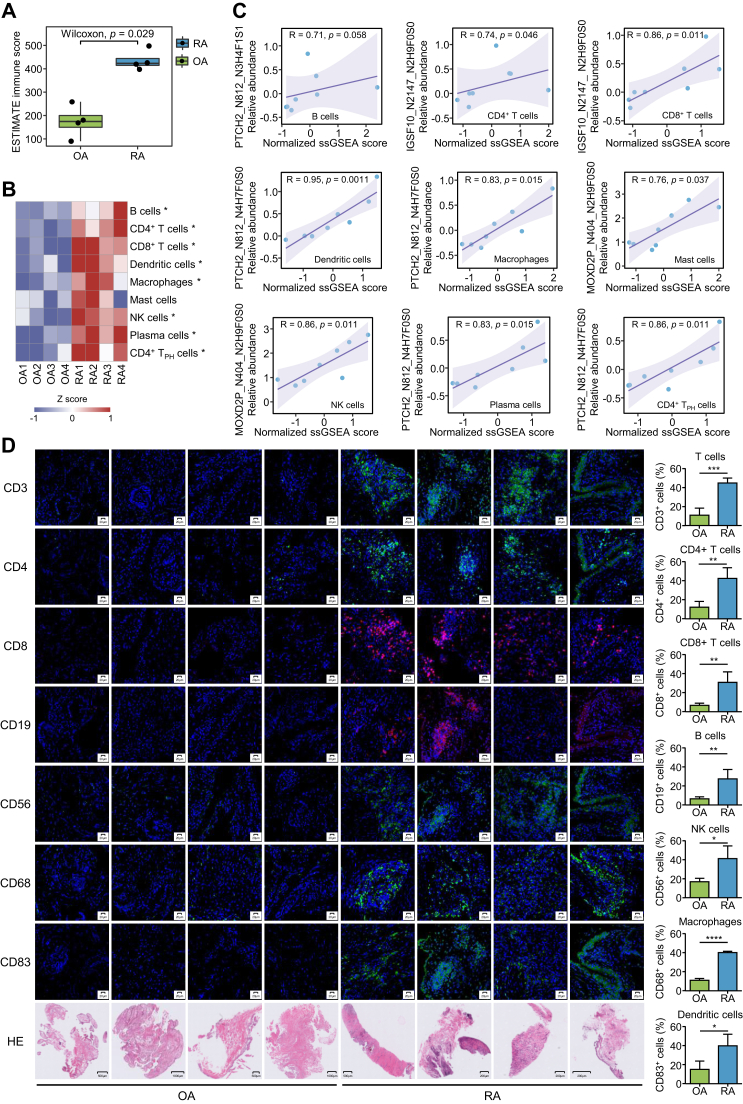
Fig. 6.
Integrative multiomics analyses to identify immune-associated N-glycopeptides in RA.A, comparison of estimated immune scores between the RA and OA groups using the ESTIMATE algorithm based on the proteomics data. The box represents the median (thick line) and the quartiles (line). B, heatmap for row-scaled enrichment scores of nine immune cell types in synovial tissues. The enrichment scores were calculated by ssGSEA using specific gene sets for these immune cell types. Rows represent infiltrating immune cells and columns represent samples (Wilcoxon rank-sum test, ∗p < 0.05). C, representative differentially expressed N-glycopeptides show a significant linear correlation with the infiltration of immune cells. R represents the regression coefficient and the p value represents the significance of the linear correlation. D, H&E staining of synovial tissue sections and immunofluorescence staining of CD3, CD4, CD8, CD19, CD56, CD68, CD83, and the percentage of CD3, CD4, CD8, CD19, CD56, and CD68, CD83-positive cells in synovial tissues from OA (n = 4) and RA (n = 4) patients. The statistics of immune-positive cells were calculated using Fiji/ImageJ. Each value represents the average obtained from four independent experiments. Data are presented as the mean ± SD. (Student’s t test, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001). H&E staining of synovial tissue sections was used to observe biopsy pathological morphology. ESTIMATE, Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data; OA, osteoarthritis; RA, rheumatoid arthritis; ssGSEA, single sample gene set enrichment analysis.