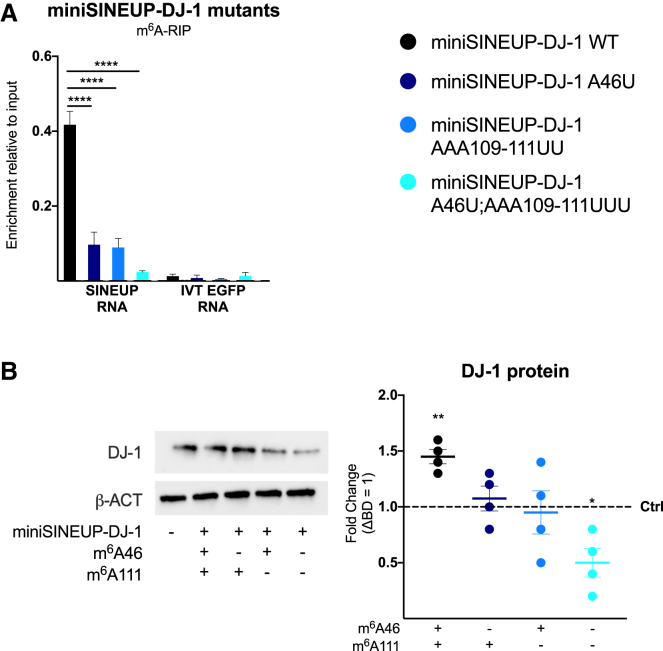
Figure 4.
m6A methylation sites regulate miniSINEUP activity
miniSINEUP-DJ-1 mutants were designed to analyze the effect of m6A sites’ loss on SINEUP activity. miniSINEUP-DJ-1 WT and its mutants A46U, AAA109-111UUU, and A46U; AAA109-111UUU were transfected in A549 cells. Presence of m6A sites in each construct is reported on the x axis. ΔBD (miniSINEUP-DJ-1 deprived of BD) was used as a negative control. (A) m6A-RIP qPCR analysis. A549 cells were transfected with miniSINEUP-DJ-1 WT and A46U, AAA109-111UUU, A46U; AAA109-111UUU mutants. Eluates from IgG immunoprecipitation were used as negative controls. IVT EGFP mRNA was spiked in total RNA extract to assess the specificity of the immunoprecipitation reaction. Data are mean ± SEM and are relative to n = 3 independent experiments. p values are calculated using two-way ANOVA and Dunnett’s multiple-comparisons test. ∗∗∗∗p < 0.0001. (B) Protein expression analysis. miniSINEUP-DJ-1 mutants’ activity was assessed using western blotting with anti-DJ-1 antibodies A549 cells. ΔBD was used as a negative control. One representative experiment is shown (left). SINEUP activity was calculated as an increase in protein quantities relative to the negative control (ΔBD). First, band intensity was normalized to the relative β-actin band. Then, fold change values were calculated normalizing on negative control-transfected cells (dotted line). On the right, data indicate single replicate values and mean ± SEM and are relative to n = 4 independent experiments. p values are calculated using unpaired t test with Mann-Whitney correction. ∗p < 0.05, ∗∗p < 0.01