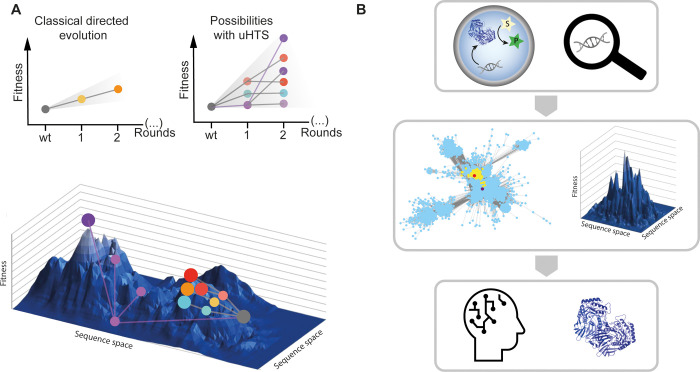
Figure 19.
Perspectives. The impact of ultrahigh-throughput screening on directed evolution. (A) Classical directed evolution constrains the campaign to the most improving variants after each round. This can yield highly improved variants in a very economical fashion but restricts the exploration of sequence space to one trajectory. With uHTS, multiple trajectories can be explored in an unbiased manner, also allowing rounds with less stringent screening regimes, increasing the likelihood of encountering synergistic effects or one-in-a-million events. (B) Droplet-based ultrahigh-throughput screening and characterization allows functional annotation of sequence space (left). Sequence similarity network from Neun et al.196 showing a novel bridgehead for functional annotation of GH3 β-glucoronidases (red). An already annotated/characterized GH3 β-glucoronidase is shown in purple while sequences directly connected to the novel bridgehead are shown in yellow. Blue sequences show all significant search hits from a MGnify query. Using ultrahigh-throughput screening coupled to high-throughput sequencing, the effect of mutations on an enzyme can be characterized on a large scale (right). Combined, we envision this large-scale sequence–function mapping to provide data for the next generation of AI-based enzyme discovery and engineering efforts.