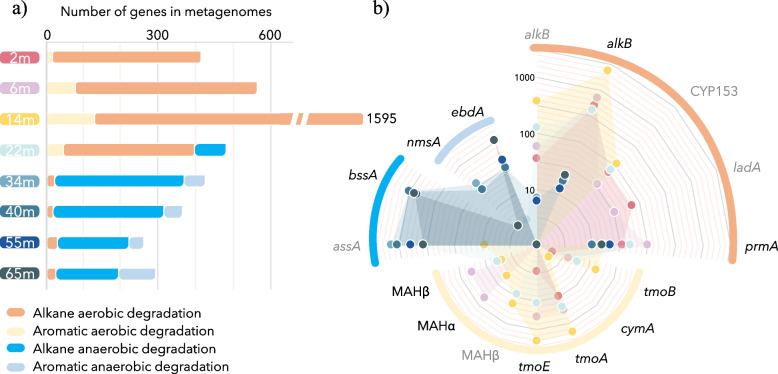
Fig. 3.
Metabolic potential for hydrocarbon degradation in Lake A. a Number of genes involved in hydrocarbon degradation in the metagenomes throughout the water column. Alkane aerobic degradation sums the number of genes of alkB, CYP153, ladA, and prmA. Aromatic aerobic degradation sums the number of genes of tmoABE, cymA, and MAHαβ. Alkane anaerobic degradation is the mean of assA and bssA gene numbers. Aromatic anaerobic degradation sums the number of genes of nmsA and ebdA. b Relative abundance of each hydrocarbon-degrading genes identified in Lake A. Color of the dots and shades represents the water depth as in a. Genes labeled in gray (assA, alkB, CYP153, ladA, and MAHβ) were identified using custom HMM profiles as described in Khot et al. [11], whereas genes labeled in black were identified using IMG-MR pipeline. Scale is logarithmic