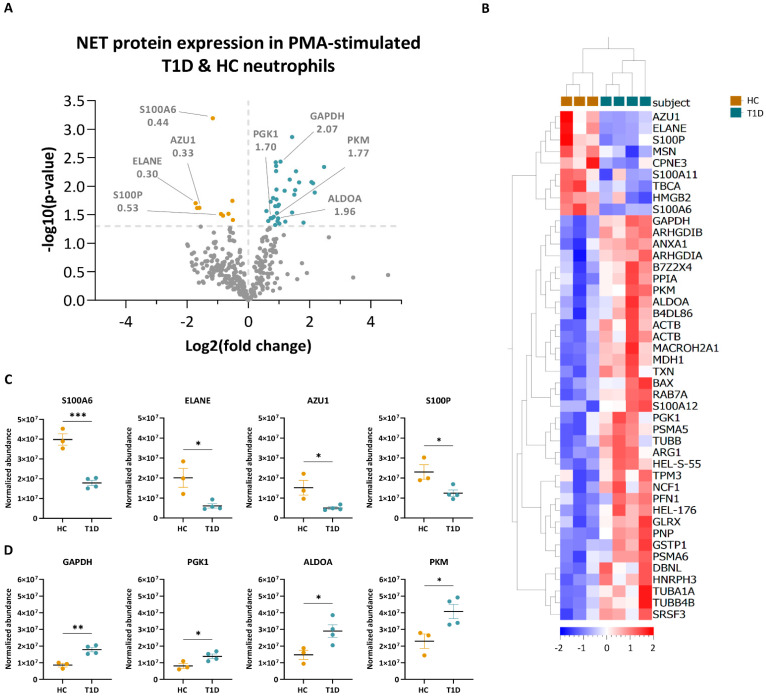
Figure 5.
Quantification of NET proteins from PMA-stimulated HC and T1D neutrophils. (A) Volcano plot based on fold change (log2) and p-value (−log10) of all differentially expressed NET proteins identified in NETing neutrophils of HC (n = 3) and T1D (n = 4) subjects in response to PMA (<1.0% FDR). Th threshold (grey dotted line) was set at p-value < 0.05. Proteins below the threshold are in grey, while proteins with higher expression levels in T1D subjects are in blue, and those in HC are in yellow. Proteins of interest, as well as their fold change values, are indicated. (B) Hierarchical clustering of normalized abundance values (Progenesis) of significantly different NET proteins of HC (yellow) and T1D (blue) subjects after PMA stimulation (<1.0% FDR, p < 0.05, unpaired two-tailed t-test). Clustering was performed on subjects (columns) and proteins (rows). Scale bar represents the normalized abundance values. (C,D) Normalized abundance values of NET proteins enriched in HC (C) and T1D (D) neutrophils in response to PMA. Abbreviations: S100A6: protein S100-A6; ELANE: neutrophil elastase; AZU1: azurocidin; S100P: protein S100-P; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; PGK1: phosphoglycerate kinase; ALDOA: fructose-bisphosphate aldolase A; PKM: pyruvate kinase. Error bars represent mean SEM (n = 3 for HC and n = 4 for T1D). Unpaired two-tailed t-test. * p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001.