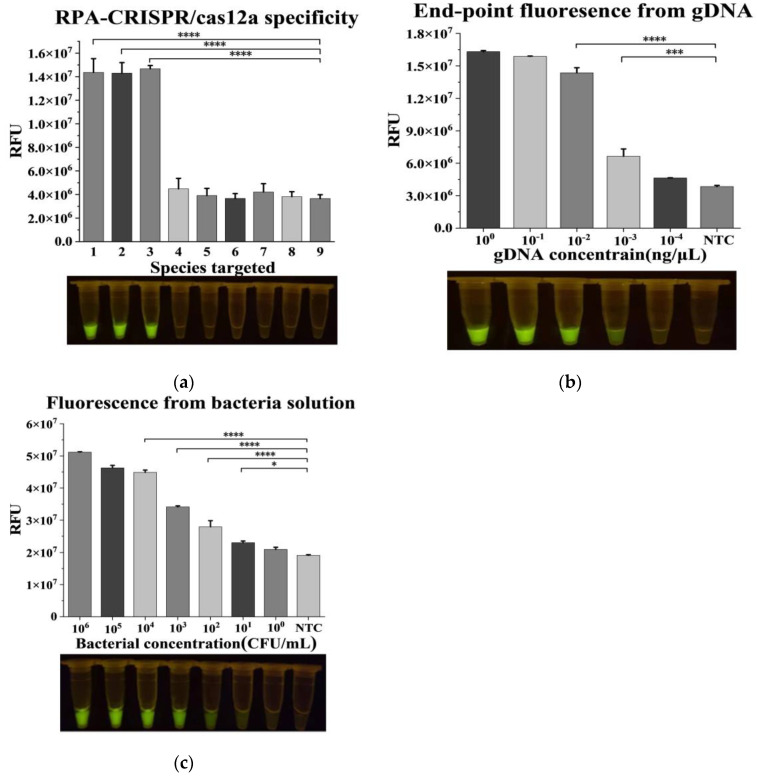
Figure 2.
Establishment and evaluation of Recombinase polymerase amplification (RPA)-CRISPR/Cas12a method. (a) Specificity analysis. Among them, the pathogenic bacteria were as follows: (1) Burkholderia Gladiolus (CICC10574), (2) Burkholderia Gladiolus (ATCC10248), (3) Burkholderia Gladiolus Toxigenic strains, (4) Salmonella, (5) Vibrio parahaemolyticus, (6) Listeria monocytogenes, (7) Bacillus cereus, (8) Staphylococcus aureus, and (9) nontarget control (use sterile water). (b) Sensitivity analysis at genomic DNA (gDNA) level. (c) Sensitivity analysis for bacterial quantity. n = 3 biological replicates, error bars represent the standard deviation of three replicates. One-way ANOVA; Tukey–Kramer was used for post hoc comparisons; * p < 0.05, *** p < 0.001, and **** p < 0.0001. NTC: nontarget control. RFU: relative fluorescence unit.