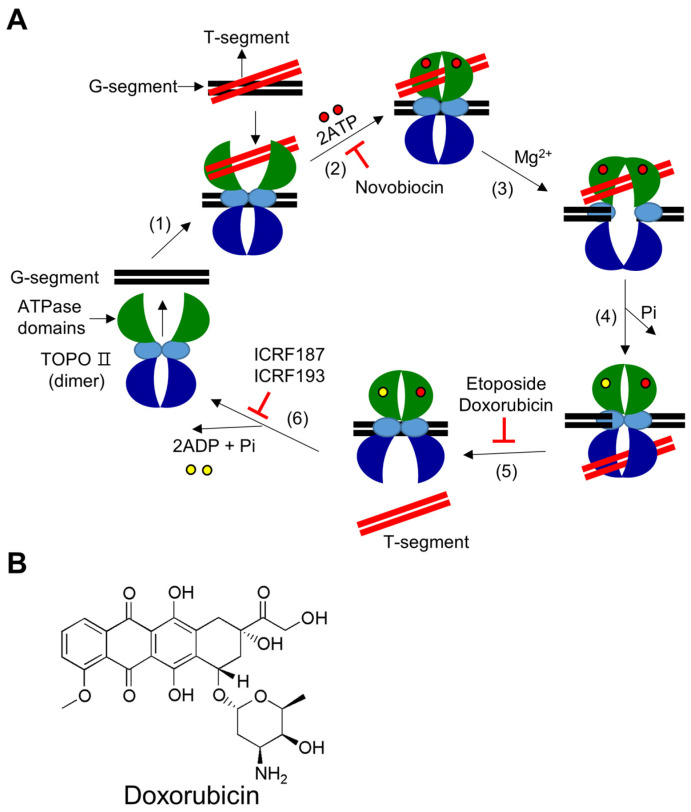
Figure 2.
Catalytic mechanism of type II DNA TOPOs and chemical structure of doxorubicin. (A) (1) DNA binding: The enzyme’s homo-dimer preferentially binds to catenated, knotted, and supercoiled DNA segments. The segment of double-stranded DNA that is cleaved during the enzymatic reaction cycle is referred to as the “G segment” (with “G” for gate), and the segment of double-stranded DNA that passes through the cleaved G segment is referred to as the “T segment” (with “T” for transported). The enzyme binds to the G segment and then to the T segment. (2) ATP binding: The binding of two ATP molecules in the ATPase domains alters the conformation of the ATPase domains from an open to a closed state. Novobiocin prevents ATP binding. (3) DNA cleavage: In the presence of Mg2+ ions, the enzyme temporarily cleaves the G segment of DNA by initiating a nucleophilic attack and forming two 5′-phosphotyrosyl bonds with the DNA backbone. (4) Strand passage: After the G segment is cleaved, the T segment is threaded through it. (5) T segment release and re-ligation: Once the T segment has passed through, it is released from the enzyme, and the cleaved G segment is rejoined. Etoposide and doxorubicin prevent the rejoining process. (6) G segment releases when the ATPase domain is opened: After the T segment is released, the enzyme stays in a closed clamp shape. Hydrolysis of ATP causes the closed clamp to open, allowing the G segment to be released and preparing the enzyme for the next reaction cycle. Bisdioxopiperazines, such as ICRF187 and ICRF193, inhibit the ATPase activity of the enzyme. (B) Chemical structural formula of doxorubicin.