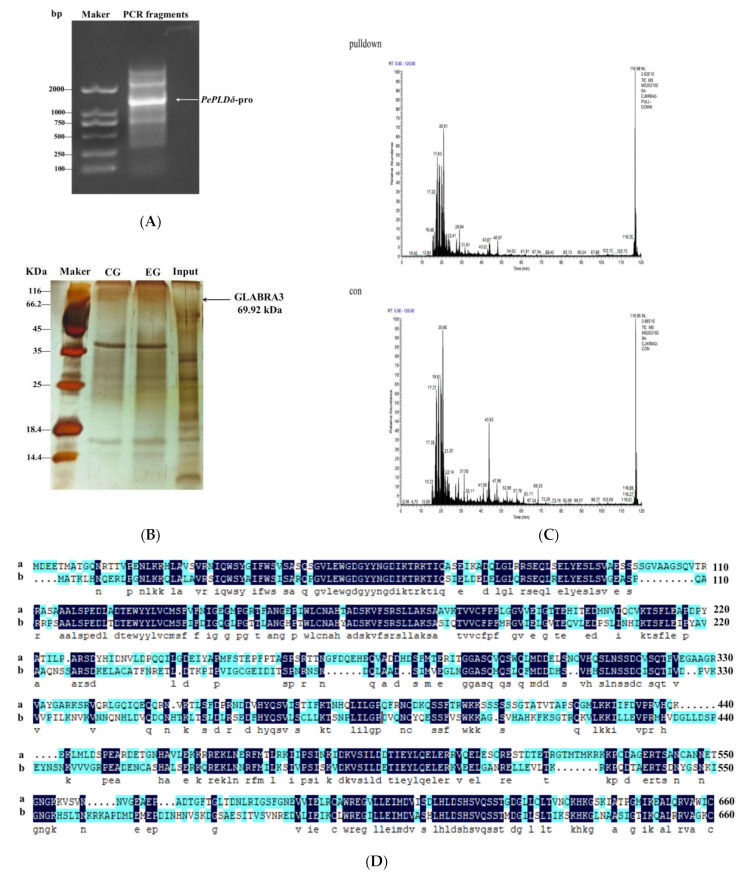
Figure 4.
DNA pull-down and mass spectrometry analysis. (A) PCR amplification of the PePLDδ probe. The PePLDδ promoter fragment was amplified by PCR, cloned, and sequenced. The probes were biotin-labeled and purified with a gel extraction kit and used for DNA pull-down. (B) DNA pull-down-enriched proteins. The biotin-labeled DNA and nucleic acid were incubated with beaver magnetic beads to form a DNA magnetic bead complex. Then, the DNA magnetic bead complex was incubated with protein extracts to form the protein-DNA-magnetic bead complex. The DNA pull-down-enriched proteins were separated using SDS-PAGE denaturing polyacrylamide gel electrophoresis and stained with a silver staining kit (Sigma-Aldrich). CG: control group; EG: experimental group. (C) Mass spectrometry analysis of DNA pull-down-enriched proteins. The green marks represent the matching isotope peaks. (D) The amino acid sequence identified by DNA pull-down covered and matched with that of PeGLABRA3; a, the protein sequence identified by DNA pull-down; b, the PeGLABRA3 protein sequence. Black shading indicates identical amino acid residues and blue shadings indicate conserved amino acids, respectively. The obtained protein sample was analyzed with mass spectrometry (Ultraflex III mass spectrometer, Bruker, Germany). The database used for protein determination was Uniprot_Arabidopsis thaliana (https://www.uniprot.org/uniprotkb?query=Arabidopsis%20thaliana, accessed on 13 August 2020), Uniprot_Populus euphratica (https://www.uniprot.org/uniprotkb?query=Populus%20euphratica, accessed on 16 December 2020), and Uniprot_Populus trichocarpa (https://www.uniprot.org/uniprotkb?query=Populus%20trichocarpa, accessed on 16 December 2020). By comparison of protein type and amino acid sequence available in the NCBI database, Arabidopsis GLABRA3 was most similar to the obtained protein, and the basic helix–loop–helix (bHLH) transcription factor GLABRA3 (GL3) was identified as a target transcription factor for binding the promoter region of PePLDδ. Con: control group.