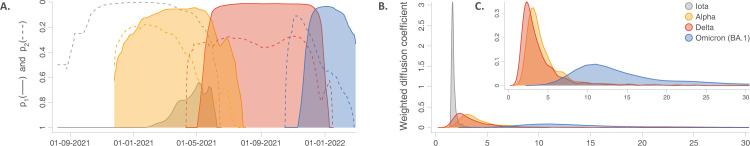
Fig 3. Comparing the introduction and dispersal dynamics of SARS-CoV-2 variants in the NYC area.
A: Evolution of (i) the probability p1 that two circulating lineages drawn at random belong to the same clade/cluster introduced into the study area (solid curves), and (ii) the proportion p2 computed as the ratio between the number of circulating clusters and the number of phylogenetic branches occurring at the same time across the study area (dashed curves). B: Posterior distributions of the weighted diffusion coefficient (km2/day) estimated for each variant. C: Posterior distributions of the weighted diffusion coefficient estimated for each variant except Iota, which allows a focus on the results obtained for the three other variants. All the results reported in Fig 3 are based on 900 trees sampled from each posterior distribution obtained by discrete (A) or continuous (B-C) phylogeographic inference.