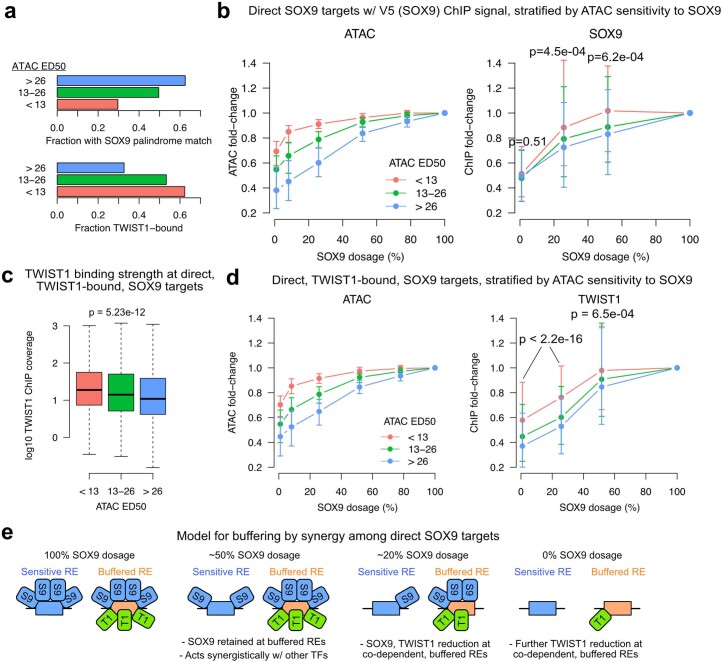
Extended Data Fig. 6. Binding of SOX9 and TWIST1 in response to SOX9 dosage changes.
(a) Fractions of rapidly downregulated REs with SOX9 palindrome match (top) or TWIST1 binding (bottom), stratified by ATAC-seq sensitivity (ED50) to SOX9 dosage (colors). N of groups by color: red, 823; green, 2,473; blue, 5983. (b) Distributions of ATAC-seq (left) or SOX9 (V5) ChIP-seq (right) fold-changes vs full SOX9 dosage at each concentration for rapidly downregulated REs with SOX9-dependent ChIP-seq signal (DESeq2 log2FoldChange < 0 in 0 vs 100 SOX9 dosage), stratified based on ATAC-seq sensitivity to SOX9 dosage (colors). N of groups by color: red, 106; green, 391; blue, 666. P-values from two-sided Kruskal-Wallis tests comparing each instance of three groups. (c) Unperturbed TWIST1 ChIP-seq signal at TWIST1-bound, rapidly downregulated REs stratified based on ATAC-seq sensitivity to SOX9 dosage (colors). N of groups by color: red, 513; green, 1318; blue, 1956. Boxplot center represents median, box bounds represent 25th and 75th percentiles, and whiskers represent 5th and 95th percentiles. (d) TWIST1 ChIP-seq fold-changes vs full SOX9 dosage at same groups of REs as in (c). In (b) and (d), points and error bars represent median and 25th and 75th percentiles of distribution. (e) Models for RE buffering by synergistic TF functions. P-values from two-sided Kruskal-Wallis tests comparing each instance of three groups.